We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 820
From Proteopedia
(Difference between revisions)
m |
|||
| Line 19: | Line 19: | ||
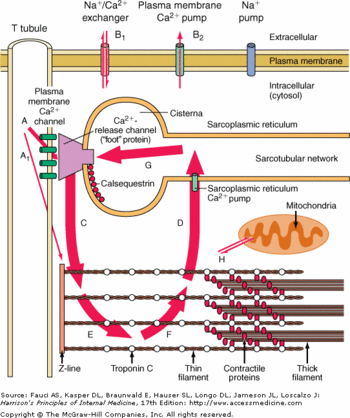
Each monomer is divided in <scene name='56/568018/Monomer_structure/5'>3 thioredoxin domains (TRX)</scene>: <scene name='56/568018/Monomer_structure/7'>the N-term</scene>, <scene name='56/568018/Monomer_structure/6'>the middle</scene> and the <scene name='56/568018/Monomer_structure/8'>C-term</scene> domains. Each of these has a regular structure: a <scene name='56/568018/Beta_sheet/4'>5 strands beta sheet core</scene> surrounded by <scene name='56/568018/Alpha_helix/3'>4 alpha helices</scene>. | Each monomer is divided in <scene name='56/568018/Monomer_structure/5'>3 thioredoxin domains (TRX)</scene>: <scene name='56/568018/Monomer_structure/7'>the N-term</scene>, <scene name='56/568018/Monomer_structure/6'>the middle</scene> and the <scene name='56/568018/Monomer_structure/8'>C-term</scene> domains. Each of these has a regular structure: a <scene name='56/568018/Beta_sheet/4'>5 strands beta sheet core</scene> surrounded by <scene name='56/568018/Alpha_helix/3'>4 alpha helices</scene>. | ||
| - | Usually these domains are involved in redox phenomena, which lead to disulfide bounds creation. Here these domains are inactive but play an important role in the polymerization of CASQ2. | + | Usually these domains are involved in redox phenomena, which lead to disulfide bounds creation. Here these domains are inactive but play an important role in the polymerization of CASQ2.<ref name="Monomère structure">http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=239372&querygi=429544235&aln=1,227,0,109</ref> |
=== Polymer Structure === | === Polymer Structure === | ||
Revision as of 17:12, 2 January 2014
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Cerrone M, Napolitano C, Priori SG. Catecholaminergic polymorphic ventricular tachycardia: A paradigm to understand mechanisms of arrhythmias associated to impaired Ca(2+) regulation. Heart Rhythm. 2009 Nov;6(11):1652-9. doi: 10.1016/j.hrthm.2009.06.033. Epub 2009 , Jun 30. PMID:19879546 doi:http://dx.doi.org/10.1016/j.hrthm.2009.06.033
- ↑ NCBI Ressource: CASQ2 calsequestrin 2 http://www.ncbi.nlm.nih.gov/gene/845
- ↑ http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=239372&querygi=429544235&aln=1,227,0,109
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Crystal Structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum (Wang et al., 1998) http://www.nature.com/nsmb/journal/v5/n6/abs/nsb0698-476.html
- ↑ The asp-rich region at the carboxyl-terminus of calsequestrin binds to Ca2+ and interacts with triadin (Shin et al., 2000) http://www.sciencedirect.com/science/article/pii/S0014579300022468
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 Beard NA, Laver DR, Dulhunty AF. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog Biophys Mol Biol. 2004 May;85(1):33-69. PMID:15050380 doi:http://dx.doi.org/10.1016/j.pbiomolbio.2003.07.001
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Beard NA, Casarotto MG, Wei L, Varsanyi M, Laver DR, Dulhunty AF. Regulation of ryanodine receptors by calsequestrin: effect of high luminal Ca2+ and phosphorylation. Biophys J. 2005 May;88(5):3444-54. Epub 2005 Feb 24. PMID:15731387 doi:http://dx.doi.org/10.1529/biophysj.104.051441