This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 823
From Proteopedia
| Line 53: | Line 53: | ||
| - | The early endosomal SNARE complex forms a four-helix bundle with a '''left-handed superhelical twist'''. The position of the Qa-, Qb-, Qc-, and R-SNAREs is the same in the neuronal and late endosomal complexes.<ref name="sutton" /><ref name="anto" /> As mentioned above, an unconventional layer of hydrophilic residues – the “0”-layer – constituted of three glutamines and one arginine, characterizes the center of all SNARE complexes. However, in vti1a an '''aspartate residue''' substitutes the glutamine. The aspartate occupies the '''same position''' as the glutamine in the neuronal and late endosomal complexes, and is involved in a '''similar organization of hydrogen bonds''' with the other layer of amino acids. Moreover, in vti1a, <scene name='56/568021/Asp_156/2'>D156</scene> of the ‘0' layer also interacts with <scene name='56/568021/Asn_152/1'>N152</scene> at the surface of the vti1a helix and with a '''water molecule'''. Also, '' | + | The early endosomal SNARE complex forms a four-helix bundle with a '''left-handed superhelical twist'''. The position of the Qa-, Qb-, Qc-, and R-SNAREs is the same in the neuronal and late endosomal complexes.<ref name="sutton" /><ref name="anto" /> As mentioned above, an unconventional layer of hydrophilic residues – the “0”-layer – constituted of three glutamines and one arginine, characterizes the center of all SNARE complexes. However, in vti1a an '''aspartate residue''' substitutes the glutamine. The aspartate occupies the '''same position''' as the glutamine in the neuronal and late endosomal complexes, and is involved in a '''similar organization of hydrogen bonds''' with the other layer of amino acids. Moreover, in vti1a, <scene name='56/568021/Asp_156/2'>D156</scene> of the ‘0' layer also interacts with <scene name='56/568021/Asn_152/1'>N152</scene> at the surface of the vti1a helix and with a '''water molecule'''. Also, <scene name='56/568021/Gln_197/1'>sx6 Q197> interacts with the '''backbone of vti1a D156''' and with '''sx6 E193''' that in turn interacts with '''vti1a R157'''. Concerning '''Vti1a R157''', it interacts with '''sx6 E196'''. This network of interactions resembles that observed in the neuronal complex.<ref> PMID: 12496247 </ref> The structure of the other layers is '''generally similar''' to that of the neuronal and late endosomal complexes. Indeed, slightly differences can be noted: in layer 6, the Qc-SNARE syntaxin 6 contains a valine, whereas the late endosomal Qc-SNARE syntaxin 8 contains a glutamate, which is involved in interactions with surface residues. Such interactions are absent in the early endosomal and the neuronal complexes. |
Regarding the SNARE motifs, they are not only connected by the central interacting layers, but also '''by surface interactions''' that often involve side chains with complementary charges. As an example, an interaction between the helices of '''syntaxin 6''' and '''vti1a''' involves a '''hydrogen bond''' between '''E143 (vti1a)''' and '''S179 (sx6)'''. At a similar position in the late endosomal complex, a '''salt bridge''' is observed between '''D157 (vti1b)''' and '''R164 (sx8)'''.<ref name="anto" /> It has to be noted that all these side chains are '''highly conserved''' between the respective SNAREs of ''Drosophila'' and mammals. Many other surface interactions connect the SNARE motifs. However, the role of these surface interactions in the stability of the whole complex is still unknown. | Regarding the SNARE motifs, they are not only connected by the central interacting layers, but also '''by surface interactions''' that often involve side chains with complementary charges. As an example, an interaction between the helices of '''syntaxin 6''' and '''vti1a''' involves a '''hydrogen bond''' between '''E143 (vti1a)''' and '''S179 (sx6)'''. At a similar position in the late endosomal complex, a '''salt bridge''' is observed between '''D157 (vti1b)''' and '''R164 (sx8)'''.<ref name="anto" /> It has to be noted that all these side chains are '''highly conserved''' between the respective SNAREs of ''Drosophila'' and mammals. Many other surface interactions connect the SNARE motifs. However, the role of these surface interactions in the stability of the whole complex is still unknown. | ||
Revision as of 19:40, 6 January 2014
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Contents |
Introduction
Important biological processes, such as synaptic transmission and cellular trafficking in Eukaryotes require SNARE proteins that are thought to play a crucial role in membrane fusion.[1][2][3][4] To connect membranes and allow their fusion, SNARE (soluble N-ethylmaleimide-sensitive-factor attachment protein receptor) proteins assemble into a core-complex of four parallel helices.[5][6][7] The SNARE complex assembly is mediated by a conserved SNARE motif consisting of 60-70 amino acids. [8] SNAREs can be divided into two categories: the v-(vesicle) SNAREs, which are found in the vesicle membrane and the t-(target) SNAREs, which are anchored in the target membrane.[9]
The best-studied SNAREs are the neuronal and the early endosomal SNARE complexes. The Neuronal SNARE complex mediates exocytosis of synaptic vesicles in the neurons and includes the vesicle protein synaptobrevin (also called VAMP), the membrane proteins SNAP-25 and syntaxin 1.[7] The early endosomal SNARE complex includes syntaxin 6, syntaxin 13, vti1a and VAMP4 and is responsible for homotypic fusion of early endosomes.[10] It has been shown that the crystal structure of the early endosomal SNARE complex resembles that of the neuronal and late endosomal complexes, but differs in surface side-chain interactions.
Membrane fusion mechanism
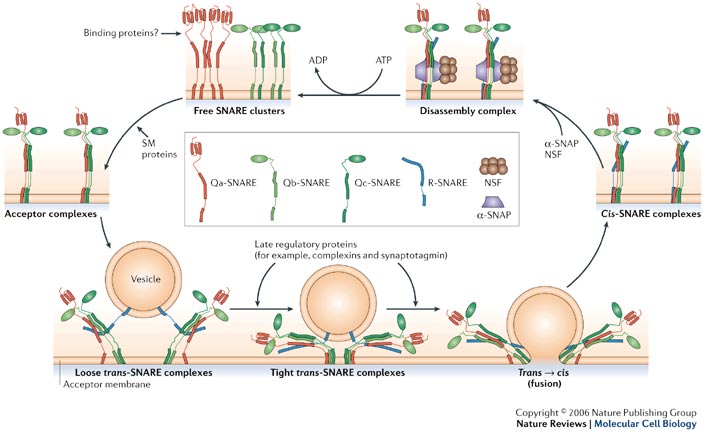
Membrane Fusion requires the assembly of the core complex. Free t-SNAREs that are organized in clusters first assemble into acceptor complexes thanks to SM (Sec1/Munc18-related) proteins. Acceptor complexes can then interact with the v-SNAREs through the N-terminal domain of the SNARE motif. This enables the formation of four-helical trans-complexes, in which only the N-terminal portions of the SNARE motifs are bound. This binding evolves from a loose to a tight state, thus leading to the formation of a fusion pore. During the fusion, the conformation relaxes to a cis-configuration. Cis-complexes dissociate thanks to proteins and cofactors (SNAPs). T- and v-SNAREs can be separated and recycled.[11]
Structure
SNARE domain
The SNARE domain is approximately 60-70 residues long and is located immediately adjacent to a C-terminal transmembrane anchor. It contains a repeating heptad pattern of hydrophobic residues. Their orientation places them in an alpha-helical structure so that all the hydrophobic side chains are located on the same face of the helix.[5][12][7] SNARE domains allow the four SNAREs protein to assemble into parallel four-helix bundles. This parallel arrangement brings the transmembrane anchors and the membranes closer. [13]
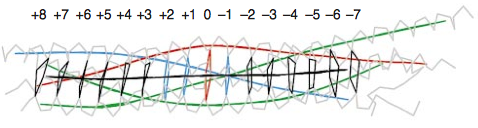
“0”-layer
The centre of the four-helix bundle is constituted of 16 layers. These layers are composed of , which are perpendicular to the axis of the four-helix bundle, except for the central “0”-layer. This last one consists of three glutamine (Q) and one arginine (R) highly conserved residues. Those highly conserved residues have led to a new classification of SNAREs into Q- and R-SNAREs. [14] Almost all membrane fusion reactions require one R-SNARE and three Q-SNAREs: Qa, Qb and Qc.[15][16] In many cases, the R-SNARE is in the vesicle, and the three Q-SNAREs are in the target membrane. For the early endosomal SNARE complex, syntaxin 13, vti1a, syntaxin 6, and VAMP4 were respectively classified as Qa-, Qb-, Qc- and R-SNAREs.
Structure of the early endosomal SNARE complex 2NPS
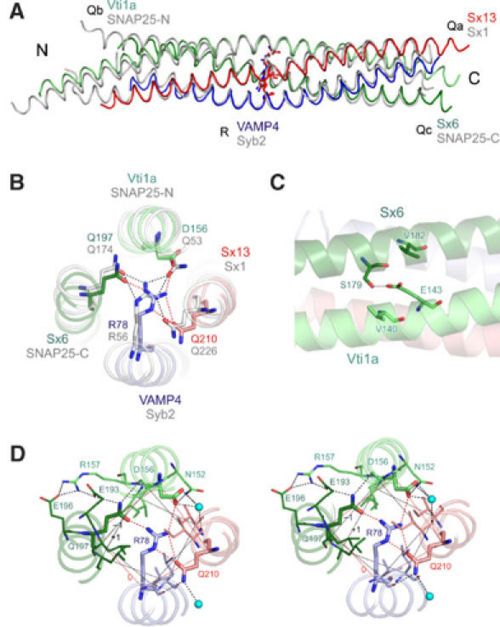
The early endosomal SNARE complex forms a four-helix bundle with a left-handed superhelical twist. The position of the Qa-, Qb-, Qc-, and R-SNAREs is the same in the neuronal and late endosomal complexes.[7][5] As mentioned above, an unconventional layer of hydrophilic residues – the “0”-layer – constituted of three glutamines and one arginine, characterizes the center of all SNARE complexes. However, in vti1a an aspartate residue substitutes the glutamine. The aspartate occupies the same position as the glutamine in the neuronal and late endosomal complexes, and is involved in a similar organization of hydrogen bonds with the other layer of amino acids. Moreover, in vti1a, of the ‘0' layer also interacts with at the surface of the vti1a helix and with a water molecule. Also,