Sandbox Reserved 830
From Proteopedia
| Line 21: | Line 21: | ||
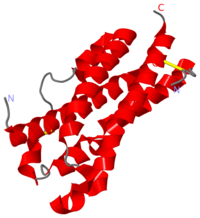
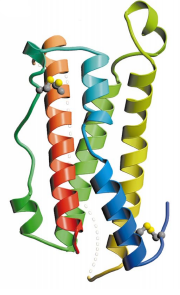
| - | Helices A and C have breaks in the hydrogen-bonding pattern of their structure, forming tight substitute hydrogen bonds with water molecules. Indeed, it results in a <scene name='56/568028/Kink_helixa/1' | + | Helices A and C have breaks in the hydrogen-bonding pattern of their structure, forming tight substitute hydrogen bonds with water molecules. Indeed, it results in a kink in helix A (<scene name='56/568028/Kink_helixa/1'and slightly in helix C between residues Gln112 and Pro116</scene>) induced by a disruption in the helical conformation, due to the Gln25 and Leu30 hydrogen bonds with four water molecules. Helix A residues between Thr27 and Ile37 take on a 310 helix conformation. With this curved structure, helices A and C enhance the compaction of the A-D and B-C parallel helix pairs, causing the core of OSM to be isolated from the solvent. |
This core is composed of two aromatic stacking groups, Phe56, Tyr, 173, Phe169 and Phe176 on one hand, and Phe170, Phe185 and Trp187 on the other hand. All these aromatic residues belong to helix D, except Phe56 (AB loop) and Phe70 (Helix B), highlighting the hydrophobicity of helix D. | This core is composed of two aromatic stacking groups, Phe56, Tyr, 173, Phe169 and Phe176 on one hand, and Phe170, Phe185 and Trp187 on the other hand. All these aromatic residues belong to helix D, except Phe56 (AB loop) and Phe70 (Helix B), highlighting the hydrophobicity of helix D. | ||
Revision as of 14:19, 9 January 2014
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Oncostatin M, also called OSM, is encoded by the OSM gene and it is mostly produced in the end of the activation of macrophages and T cells. OSM belongs to the family of gp130 cytokines implying that it signals through the receptors containing gp130. OSM has been shown to have a lot of pleiotropic functions in cell proliferation, differentiation and inflammatory response. Thus, studies highlight its roles in cancer, bone and liver metabolism alteration, as well as in severe inflammatory disease, such as lung and skin inflammatory disease, atherosclerosis, cardiovascular diseases, and rheumatoid polyarthritis.
Human Oncostatin M
Structure
OSM is a compact molecule with dimensions of approximately 20 Å x 27 Å x 56 Å, that fit with the up-up-down-down structure (Fig.1).
OSM structure is composed of the four main α helical region (helix A, residues 10–37; helix B, residues 67–90; helix C, residues 105–131; helix D, residues 159–185) linked by two long overhand loops (AB loop, residues 38–66; CD loop, residues 130–158) and one short loop (BC loop, residues 91–104). Globally, OSM arrangement corresponds to .
Helices A and C have breaks in the hydrogen-bonding pattern of their structure, forming tight substitute hydrogen bonds with water molecules. Indeed, it results in a kink in helix A (