We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Brr2
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
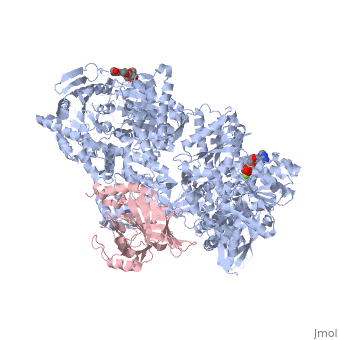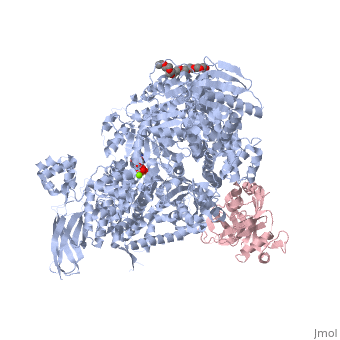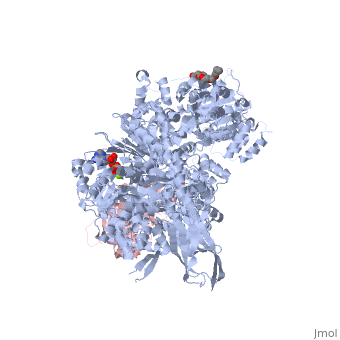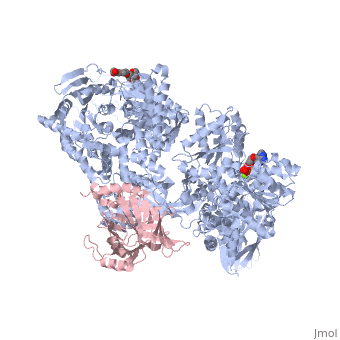
| - | <StructureSection load=' | + | <StructureSection load='4bgd' size='450' side='right' scene='' caption='Structure of yeast Brr2 complex with pre-mRNA-splicing factor 8 (PDB code [[4bgd]])'> |
='''Structure of Brr2'''= | ='''Structure of Brr2'''= | ||
| Line 40: | Line 40: | ||
*[http://www.rcsb.org/pdb/explore/explore.do?structureId=2P6U Apo structure of the Hel308 superfamily 2 helicase, in the RCSB Protein Data Bank] | *[http://www.rcsb.org/pdb/explore/explore.do?structureId=2P6U Apo structure of the Hel308 superfamily 2 helicase, in the RCSB Protein Data Bank] | ||
*[http://www.rcsb.org/pdb/explore/explore.do?structureId=2p6r Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA, in the RCSB Protein Data Bank] | *[http://www.rcsb.org/pdb/explore/explore.do?structureId=2p6r Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA, in the RCSB Protein Data Bank] | ||
| + | |||
| + | ==3D structures of Brr2== | ||
| + | |||
| + | See [[Helicase]] | ||
=References= | =References= | ||
<References/> | <References/> | ||
Revision as of 06:28, 18 August 2014
| |||||||||||
Additional Resources
- Crystal structure of the second Sec63 domain of yeast Brr2, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P41212 Form, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P212121 Form, in the RCSB Protein Data Bank
- DNA REPAIR HELICASE HEL308,in the RCSB Protein Data Bank, in the RCSB Protein Data Bank
- Apo structure of the Hel308 superfamily 2 helicase, in the RCSB Protein Data Bank
- Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA, in the RCSB Protein Data Bank
3D structures of Brr2
See Helicase
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the Spliceosome. Biochemistry. 2012 Apr 10. PMID:22471593 doi:10.1021/bi201215r
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008 Nov 12;16(11):1605-15. PMID:19000813 doi:10.1016/j.str.2008.08.011
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 5.0 5.1 5.2 Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007 Jul;14(7):647-52. Epub 2007 Jun 10. PMID:17558417 doi:10.1038/nsmb1246
- ↑ Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008 Feb 22;283(8):5118-26. Epub 2007 Dec 4. PMID:18056710 doi:10.1074/jbc.M707548200