Hexokinase
From Proteopedia
| Line 14: | Line 14: | ||
== Structure of Hexokinase == | == Structure of Hexokinase == | ||
Hexokinase is composed of an N-terminal regulatory domain and a C-terminal catalytic domain. These two domains are <scene name='Bawel_sandbox1/Hexokinase/2'>joined together by an alpha helix</scene>. The molecular weights of hexokinases are around 100 kD. Each domain weighs about 50kD and contains a <scene name='Bawel_sandbox1/Glucose_binding_site/1'>glucose binding site</scene>. But, only in hexokinase II do both halves have functional active sites. The tertiary structure of hexokinase includes an open alpha/beta sheet. There is a large amount of variation associated with this structure. The ATP-binding domain is composed of <scene name='Bawel_sandbox1/5_beta_sheets/3'>five beta sheets and two alpha helices</scene>. In this open alph/beta sheet four of the beta sheets are parallel and one is in the anitparallel directions. The alpha helices and beta loops connect the beta sheets to produce this open alpha/beta sheet. | Hexokinase is composed of an N-terminal regulatory domain and a C-terminal catalytic domain. These two domains are <scene name='Bawel_sandbox1/Hexokinase/2'>joined together by an alpha helix</scene>. The molecular weights of hexokinases are around 100 kD. Each domain weighs about 50kD and contains a <scene name='Bawel_sandbox1/Glucose_binding_site/1'>glucose binding site</scene>. But, only in hexokinase II do both halves have functional active sites. The tertiary structure of hexokinase includes an open alpha/beta sheet. There is a large amount of variation associated with this structure. The ATP-binding domain is composed of <scene name='Bawel_sandbox1/5_beta_sheets/3'>five beta sheets and two alpha helices</scene>. In this open alph/beta sheet four of the beta sheets are parallel and one is in the anitparallel directions. The alpha helices and beta loops connect the beta sheets to produce this open alpha/beta sheet. | ||
| - | |||
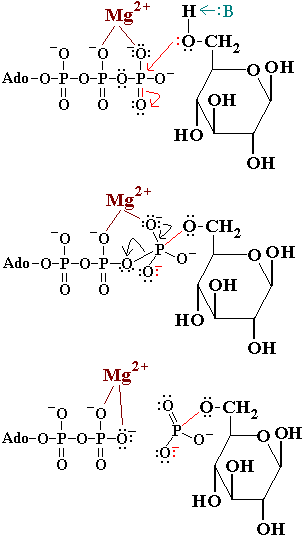
[[Image:Hexokinase_mechanism2.GIF|350px|left|thumb]] | [[Image:Hexokinase_mechanism2.GIF|350px|left|thumb]] | ||
| Line 21: | Line 20: | ||
In the first reaction of glycolysis, the gamma-phosphoryl group of an ATP molecule is transferred to the oxygen at the C-6 of glucose. Hexokinase catalyzes this phosphoryl group transfer. To start this reaction, ATP forms a complex with magnesium (II) ion and glucose binds to hexokinase. The magnesium-ATP complex then binds with the hexokinase-glucose complex and forms an intermediate (Zeng, et al. present a picture showing the interctions of brain hexokinase with ATP). <scene name='Bawel_sandbox1/Asp_532_and_thr_680/2'>Asp 532 and Thr 680</scene> are thought to be involved in binding the magnesium ion in the magnesium-ATP complex [4]. The hydroxyl group on the terminal phosphoryl group of the ATP molecule nucleophilically attacks carbon 6 on glucose. This produces glucose-6-phosphate still bound to hexokinase and ADP still in complex with magnesium ion [5]. Glucose-6-phosphate and the magnesium-ADP complex leave hexokinase. Glucose-6-phosphate and ADP are the products of this reaction. Hexokinase undergoes an induced-fit conformational change when it binds to glucose, which ultimately prevents the hydrolysis of ATP. It also experiences potent allosteric inhibition under physiological concentrations by its immediate products, glucose-6-phosphate [4]. This is a mechanism by which the influx of substrate into the glycolytic pathway is controlled. | In the first reaction of glycolysis, the gamma-phosphoryl group of an ATP molecule is transferred to the oxygen at the C-6 of glucose. Hexokinase catalyzes this phosphoryl group transfer. To start this reaction, ATP forms a complex with magnesium (II) ion and glucose binds to hexokinase. The magnesium-ATP complex then binds with the hexokinase-glucose complex and forms an intermediate (Zeng, et al. present a picture showing the interctions of brain hexokinase with ATP). <scene name='Bawel_sandbox1/Asp_532_and_thr_680/2'>Asp 532 and Thr 680</scene> are thought to be involved in binding the magnesium ion in the magnesium-ATP complex [4]. The hydroxyl group on the terminal phosphoryl group of the ATP molecule nucleophilically attacks carbon 6 on glucose. This produces glucose-6-phosphate still bound to hexokinase and ADP still in complex with magnesium ion [5]. Glucose-6-phosphate and the magnesium-ADP complex leave hexokinase. Glucose-6-phosphate and ADP are the products of this reaction. Hexokinase undergoes an induced-fit conformational change when it binds to glucose, which ultimately prevents the hydrolysis of ATP. It also experiences potent allosteric inhibition under physiological concentrations by its immediate products, glucose-6-phosphate [4]. This is a mechanism by which the influx of substrate into the glycolytic pathway is controlled. | ||
| - | |||
== Kinetics and Inhibition of Hexokinase == | == Kinetics and Inhibition of Hexokinase == | ||
| Line 29: | Line 27: | ||
G6P inhibits hexokinase by binding to the N-terminal domain(this is simple feedback inhibition). It competitively inhibits the binding of ATP [8]. If the cell is not using the G6P that it is making, then it stops making it. In this way, hexokinase can also slow down glycolysis. Hexokinase I is thought to be the "pacemaker" of glycolysis in brain tissue and red blood cells [4]. Inorganic phosphate allosterically relieves hexokinase of inhibition by G6P [8]. | G6P inhibits hexokinase by binding to the N-terminal domain(this is simple feedback inhibition). It competitively inhibits the binding of ATP [8]. If the cell is not using the G6P that it is making, then it stops making it. In this way, hexokinase can also slow down glycolysis. Hexokinase I is thought to be the "pacemaker" of glycolysis in brain tissue and red blood cells [4]. Inorganic phosphate allosterically relieves hexokinase of inhibition by G6P [8]. | ||
</StructureSection> | </StructureSection> | ||
| - | __NOTOC__ | ||
==3D structures of hexokinase== | ==3D structures of hexokinase== | ||
Revision as of 10:37, 18 August 2014
| |||||||||||
Contents |
3D structures of hexokinase
Updated on 18-August-2014
3o08, 3o1b, 3o1w, 3o4w, 3o6w, 3o80, 4jax – KlHK – Kluyveromyces lactis
2e2n – StHK – Sulfolobus tokodaii
2e2o - StHK + glucose
2e2p – StHK + ADP
2e2q - StHK + ADP + xylose + Mg
3o5b, 3o8m – KlHK + glucose
1bdg – HK + glucose – Schistosoma mansoni
Hexokinase I
3b8a – yHK I + glucose – yeast
1hkg – yHK I
1dgk – hHK I (mutant) + ADP + glucose – human
1cza - hHK I (mutant) + ADP + glucose-6-phosphate + glucose
1bg3 - HK I + glucose-6-phosphate + glucose - rat
1qha – hHK I + AMP-PNP
1hkc - hHK I + phosphate + glucose
1hkb, 4f9o - hHK I + glucose-6-phosphate + glucose
4fpa - hHK I (mutant) + glucose-6-phosphate + glucose
4foe - hHK I + mannose-6-phosphate + glucose
4foi - hHK I (mutant) + glucose 1,6-bisphosphate + glucose
4fpb - hHK I + 1,5-anhydroglucitol-6-phosphate + glucose
Hexokinase II
1ig8, 2yhx – yHK II
2nzt – hHK II
Hexokinase III
3hm8 – hHK III C terminal
Hexokinase IV (Glucokinase GK)
3qic – hHK IV residues 12-465 (mutant)
1v4t – hGK
3mcp – GK – Parabacterioides distasonis
2qm1 – GK – Enterococcus faecalis
3vgk – SgGK – Streptomyces griseus
1q18 – EcGK – Escherichia coli
4eun – GK – Janibacter
3vov – GK – Thermus thermophilus
Hexokinase IV binary complex
1sz2 - EcGK + glucose
3vgm - SgGK + glucose
3idh - hHK IV residues 12-465 + glucose
3h1v, 3imx, 3a0i, 3goi,1v4s, 3s41, 3vev, 3vf6, 4dch, 4dhy - hHK IV residues 12-465 + synthetic activator
3fr0, 4l3q, 4ise, 4isf, 4isg, 4iwv, 4ixc - hHK IV residues 12-465 + activator
Hexokinase IV ternary complex
3id8, 3fgu - hHK IV residues 12-465 + AMP-PNP + glucose
3f9m - hHK IV residues 12-465 + activator + glucose
2q2r - GK + glucose + ADP – Trypanosoma cruzi
3vgl - SgGK + glucose + AMP-PNP
3vey - hGK + glucose + ATPgS
ADP-dependent GK
1gc5 – AGK + ADP – Thermococcus litoralis
1l2l – AGK – Pyrococcus horikoshii
1ua4 - AGK – Pyrococcus furiosus
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
1.↑ Pollard-Knight D, Cornish-Bowden A. Mechanism of liver glucokinase. Mol Cell Biochem. 1982 Apr 30;44(2):71-80. PMID:7048063
2.↑ 2.0 2.1 Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004 Mar;12(3):429-38. PMID:15016359 doi:10.1016/j.str.2004.02.005
3.↑ Postic C, Shiota M, Magnuson MA. Cell-specific roles of glucokinase in glucose homeostasis. Recent Prog Horm Res. 2001;56:195-217. PMID:11237213
4.↑ Zeng C, Aleshin A, Hardie J, Harrison R, Fromm H. ATP-Binding site of Human Brain Hexokinase as Studied by Molecular Modeling and Site-Directed Mutagenesis. Biochem. 1996 Aug 6;35:13157-13164.
5.↑ hammes G, and Kochavi D. Studies of the Enzyme Hexokinase: Kinetic Inhibition by Products. Massachusetts Institute of Technology. 1961 Oct 5.
6.↑ Ralph E, Thomson J, Almaden J, Sun S. Glucose Modulation fo Glucokinase Activation by Small Molecules. Biochem. 2008 Feb 15;47:5028-5036.
7.↑ Pal P, and Miller B. Activating Mutations in the Human Glucokinase Gene Revealed by Genetic Selection. Biochem. 2008 Dec 3;48:814-816.
8.↑ Aleshin A, Malfois M, Liu X, Kim C, Fromm H, Honzatko R, Koch M, Svergun D. Nonaggregating Mutant of Recombinant Human Hexokinase I Exhibits Wild-Type Kinetics and Rod-like Conformations in Solution. Biochem. 1999 Apr 29;38:8359-8366.
Seth Bawel and Kyle_Schroering created this page in Che 361 at Wabash College.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Ann Taylor, Alexander Berchansky