Journal:BAMBEd:Acetylcholinesterase: Substrate Traffic and Inhibition
From Proteopedia
(Difference between revisions)

| Line 41: | Line 41: | ||
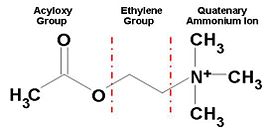
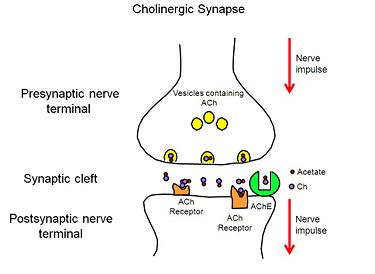
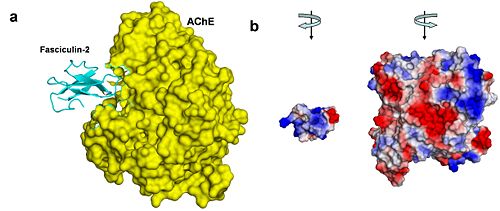
Reflected in our design are two key concepts of AChE biology: the mechanism by which AChE hydrolyses ACh (the substrate traffic story), and how the Green Mamba Snake toxin, FAS-II, inhibits the hydrolysis of ACh (the inhibition story)<ref>PMID:18586019</ref>. Two physical models were designed and fabricated using a combination of computational molecular modeling and 3D printing technology: ''Tc''AChE in complex with a modeled ACh ligand, and ''Tc''AChE in complex with FAS-II. Both models were designed using the respective protein data bank (PDB) files: 2ace for the ''Tc''AChE/ACh complex and 1fss for the''Tc''AChE/FAS-II complex, and RasMol computer modeling program. | Reflected in our design are two key concepts of AChE biology: the mechanism by which AChE hydrolyses ACh (the substrate traffic story), and how the Green Mamba Snake toxin, FAS-II, inhibits the hydrolysis of ACh (the inhibition story)<ref>PMID:18586019</ref>. Two physical models were designed and fabricated using a combination of computational molecular modeling and 3D printing technology: ''Tc''AChE in complex with a modeled ACh ligand, and ''Tc''AChE in complex with FAS-II. Both models were designed using the respective protein data bank (PDB) files: 2ace for the ''Tc''AChE/ACh complex and 1fss for the''Tc''AChE/FAS-II complex, and RasMol computer modeling program. | ||
---- | ---- | ||
| - | ===='''Features of the Substrate Traffic Story:''a Model of'' AChE/ACh'''==== | + | ===='''Features of the Substrate Traffic Story:''a Model of'' <scene name='Sandbox_250/Ache_ach/1'>AChE/ACh</scene>'''==== |
---- | ---- | ||
| - | <scene name='Sandbox_250/Ache_ach/1'>AChE/ACh</scene> | ||
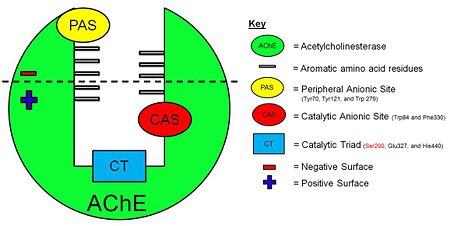
[[Image:AChE-Page-schematic-gorge.jpg|left|thumb|alt= Alt text| Figure 3. Schematic illustration of AChE. |450px]] | [[Image:AChE-Page-schematic-gorge.jpg|left|thumb|alt= Alt text| Figure 3. Schematic illustration of AChE. |450px]] | ||
{{clear}} | {{clear}} | ||
Revision as of 08:49, 26 February 2014
| |||||||||||
References
- ↑ Greenblatt HM, Dvir H, Silman I, Sussman JL. Acetylcholinesterase: a multifaceted target for structure-based drug design of anticholinesterase agents for the treatment of Alzheimer's disease. J Mol Neurosci. 2003;20(3):369-83. PMID:14501022 doi:10.1385/JMN:20:3:369
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Harel M, Kleywegt GJ, Ravelli RB, Silman I, Sussman JL. Crystal structure of an acetylcholinesterase-fasciculin complex: interaction of a three-fingered toxin from snake venom with its target. Structure. 1995 Dec 15;3(12):1355-66. PMID:8747462
- ↑ Silman I, Sussman JL. Acetylcholinesterase: how is structure related to function? Chem Biol Interact. 2008 Sep 25;175(1-3):3-10. Epub 2008 Jun 6. PMID:18586019 doi:10.1016/j.cbi.2008.05.035
- ↑ Kessel A and Ben-Tal N (Dec. 2010) Introduction to Proteins: Structure, Function, and Motion. Chapman & Hall/CRC Mathematical & Computational Biology. ISBN: 9781439810712
Acknowledgements
1. Howard Hughes Medical Institue Pre-College Program
2. Center for BioMolecular Modeling, Milwaukee School of Engineering
3. The Rockefeller University Center for Clinical and Translational Science
4. The Rockefeller University S.M.A.R.T Team Program
5. The Rockefeller University Science Outreach Program
6. Touro College of Pharmacy
7. Michal Harel, Weizmann Institute of Science
8. Natural Sciences Department,Hostos Community College, Bronx, NY
9. Malcolm Twist
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Michal Harel, Daviana Dueno, Randol Mata, Mary Acheampong, Allison Granberry, Alafia Henry, Marisa L. VanBrakle
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.