This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox reserved 915
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
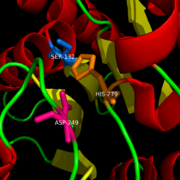
[[Image:Catalytic_triad.png|right|200|thumb|Catalytic Triad of MGL structure]] | [[Image:Catalytic_triad.png|right|200|thumb|Catalytic Triad of MGL structure]] | ||
====Binding==== | ====Binding==== | ||
| - | 2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. See Overall Reaction. | + | 2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. '''See Overall Reaction.''' |
===Inhibition=== | ===Inhibition=== | ||
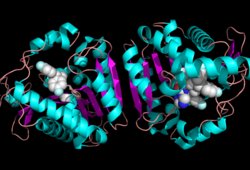
Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can not be inhibited, but it can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This natural inhibition phenomenon is known as interfacial activation. | Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can not be inhibited, but it can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This natural inhibition phenomenon is known as interfacial activation. | ||
| Line 20: | Line 20: | ||
==Overall Reaction== | ==Overall Reaction== | ||
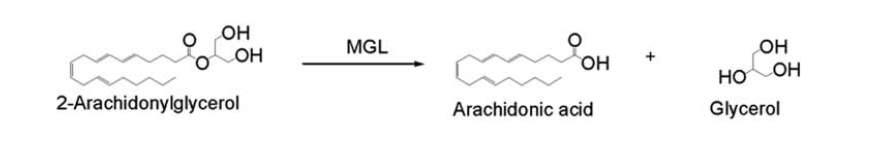
| - | In this reaction 2-AG binds to the catalytic triad in the oxyanion hole in the active site. In the oxyanion hole the oxygen of the substrate is stabilized by two nitrogen atoms during the transition step of the catalytic reaction. The triad activates the nucleophilic serine and cleaves the ester bond of 2-AG that is being stabilized by its carbonyl group that is attached to the oxyanion hole. The glycerol molecule is released and it might diffuse to the narrow "exit hole", while the arachidonic acid would diffuse back to the top of the tunnel and leave the protein. | ||
[[Image:Reaction.PNG]] | [[Image:Reaction.PNG]] | ||
| + | In this reaction 2-AG binds to the catalytic triad in the oxyanion hole in the active site. In the oxyanion hole the oxygen of the substrate is stabilized by two nitrogen atoms during the transition step of the catalytic reaction. The triad activates the nucleophilic serine and cleaves the ester bond of 2-AG that is being stabilized by its carbonyl group that is attached to the oxyanion hole. The glycerol molecule is released and it might diffuse to the narrow "exit hole", while the arachidonic acid would diffuse back to the top of the tunnel and leave the protein. | ||
== Literature == | == Literature == | ||
Revision as of 00:20, 25 March 2014
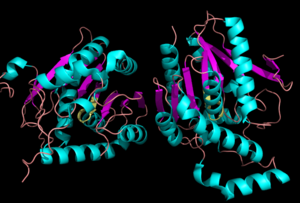
Monoglyceride Lipase (MGL)
| |||||||||||