Malate dehydrogenase
From Proteopedia
| Line 1: | Line 1: | ||
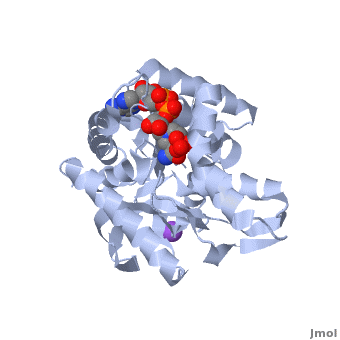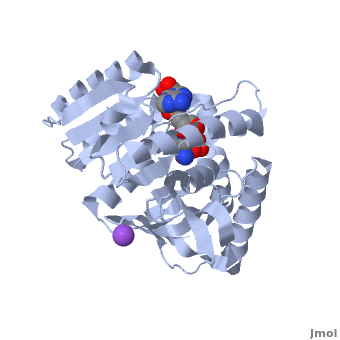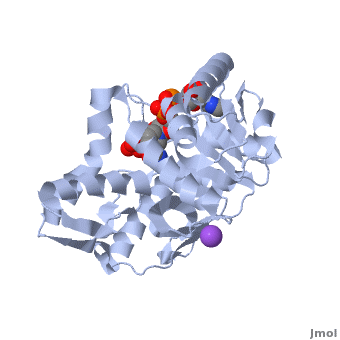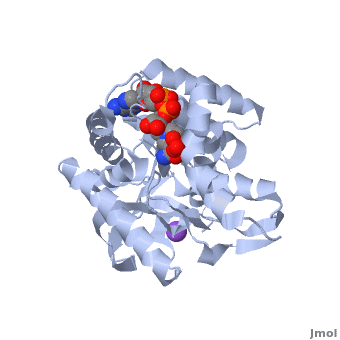
<StructureSection load='2x0i' size='450' side='right' scene='Malate_dehydrogenase/Cv/1' caption='Malate dehydrogenase complex with NAD, sulfate and Na+ ion (purple) (PDB code [[2x0i]])'> | <StructureSection load='2x0i' size='450' side='right' scene='Malate_dehydrogenase/Cv/1' caption='Malate dehydrogenase complex with NAD, sulfate and Na+ ion (purple) (PDB code [[2x0i]])'> | ||
| - | + | ||
{{Clear}} | {{Clear}} | ||
[[Malate dehydrogenase|Malate Dehydrogenase]] (MDH)(PDB entry [http://www.pdb.org/pdb/explore/explore.do?structureId=2X0I 2x0i]) is most known for its role in the metabolic pathway of the tricarboxylic acid cycle, also know as the Kreb's Cycle, which is critical to cellular respiration in cells [http://en.wikipedia.org/wiki/Citric_acid_cycle]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance <ref>PMID:12537350</ref>. It is classified as an oxidoreductase[http://en.wikipedia.org/wiki/Oxidoreductase]. Malate Dehydrogenase has been extensively studied due to its many isozymes <ref>PMID:20173310</ref>. The enzyme exists in two places inside a cell: the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; however, in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport <ref>PMID:20173310</ref>. This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer depending on the location of the enzyme and the organism it is located in <ref>PMID: 9834842</ref>. During catalysis, the enzyme subunits are non-cooperative between active sites. The mitochondrial MDH is complexly, allosterically controlled by citrate, but no other known metabolic regulation mechanisms have been discovered. Further, the exact mechanism of regulation has yet to be discovered <ref>PMID:7574693</ref>. Kinetically, the pH of optimization is 7.6 for oxaloacetate conversion and 9.6 for malate conversion. The reported K(m) value for malate conversion is 215 uM and the V(max) value is 87.8 uM/min <ref>PMID:19277715</ref>. For halophilic MDH details see [[Halophilic malate dehydrogenase]]. | [[Malate dehydrogenase|Malate Dehydrogenase]] (MDH)(PDB entry [http://www.pdb.org/pdb/explore/explore.do?structureId=2X0I 2x0i]) is most known for its role in the metabolic pathway of the tricarboxylic acid cycle, also know as the Kreb's Cycle, which is critical to cellular respiration in cells [http://en.wikipedia.org/wiki/Citric_acid_cycle]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance <ref>PMID:12537350</ref>. It is classified as an oxidoreductase[http://en.wikipedia.org/wiki/Oxidoreductase]. Malate Dehydrogenase has been extensively studied due to its many isozymes <ref>PMID:20173310</ref>. The enzyme exists in two places inside a cell: the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; however, in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport <ref>PMID:20173310</ref>. This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer depending on the location of the enzyme and the organism it is located in <ref>PMID: 9834842</ref>. During catalysis, the enzyme subunits are non-cooperative between active sites. The mitochondrial MDH is complexly, allosterically controlled by citrate, but no other known metabolic regulation mechanisms have been discovered. Further, the exact mechanism of regulation has yet to be discovered <ref>PMID:7574693</ref>. Kinetically, the pH of optimization is 7.6 for oxaloacetate conversion and 9.6 for malate conversion. The reported K(m) value for malate conversion is 215 uM and the V(max) value is 87.8 uM/min <ref>PMID:19277715</ref>. For halophilic MDH details see [[Halophilic malate dehydrogenase]]. | ||
Revision as of 07:48, 19 August 2014
| |||||||||||
3D Structures of Malate Dehydrogenase
Updated on 19-August-2014
The holo-MDH contains NAD or its derivatives while the apo-MDH lacks it.
Holo-MDH
2x0r – HmMDH (mutant)+NAD - Haloarcula marismortui
1o6z - HmMDH (mutant)+NADH
1hlp – HmMDH+NAD
1x0i – AfMDH+NADH – Archaeoglobus fulgidus
2x0j - AfMDH+etheno-NAD
1hlp – HmMDH+NAD
1x0i – AfMDH+NADH
2x0j - AfMDH+etheno-NAD
1ib6, 1ie3 – EcMDH (mutant)+NAD - Escherichia coli
1emd – EcMDH+NAD+citrate
3i0p – MDH+NAD – Entamoeba histolytica
3gvh – BmMDH+NAD – Brucella melitensis
3gvi - BmMDH+ADP
2hjr – MDH+adenosine diphosphoribose – Cryptosporidium parvum
2dfd – MDH+NAD – human type 2
1wze – TfMDH (mutant)+NAD – Thermus flavus
1wzi - TfMDH (mutant)+NDP
1bdm - TfMDH (mutant)+beta-6-hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide
1bmd – TfMDH+NAD
1y7t – TtMDH+NADPH – Thermus thermophilus
2cvq - TtMDH+NADP
1v9n – MDH+NADPH – Pyrococcus horikoshii
1z2i – MDH+NAD – Agrobacterium tumefaciens
1uxg, 1uxh, 1uxi, 1uxj, 1uxk, 1ur5 – ChaMDH (mutant)+NAD – Chloroflexus aurantiacus
1guz, 1guy, 1gv0 – CvMDH+NAD – Chlorobium vibrioforme
1civ – MDH+NADP – Flaveria bidentis
1b8u, 1b8v – AaMDH+NAD - Aquaspirillum arcticum
5mdh – SsMDH+NAD+alpha-ketomalonic acid – Sus scrofa
4mdh – SsMDH+NAD
4i1i – LmMd + NAD – Leishmania major
apo-MDH
2j5r, 2j5k, 2j5q, 1d3a, 4jco – HmMDH
2hlp – HmMDH (mutant)
3hhp, 2pwz – EcMDH
3fi9 – MDH – Porphyromonas gingivalis
3d5t - MDH – Burkholderia pseudomallei
2d4a – MDH – Aeropyrum pernix
1iz9, 4kde, 4kdf - TtMDH
1sev, 1smk – MDH – Citrullus lanatus
1gv1 – CvMDH
1b8p – AaMDH
7mdh – MDH – Sorgum bicolor
1mld – SsMDH
2cmd - EcMd+citrate
3nep – Md – Salinibacter ruber
3p7m – Md – Francisella tularensis
3tl2 – Md – Bacillus anthracis
4e0b – Md – Vibrio vulnificus
4h7p - LmMd
4bgt – CaMd
4cl3 – ChaMd
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Minarik P, Tomaskova N, Kollarova M, Antalik M. Malate dehydrogenases--structure and function. Gen Physiol Biophys. 2002 Sep;21(3):257-65. PMID:12537350
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310
- ↑ Musrati RA, Kollarova M, Mernik N, Mikulasova D. Malate dehydrogenase: distribution, function and properties. Gen Physiol Biophys. 1998 Sep;17(3):193-210. PMID:9834842
- ↑ Boernke WE, Millard CS, Stevens PW, Kakar SN, Stevens FJ, Donnelly MI. Stringency of substrate specificity of Escherichia coli malate dehydrogenase. Arch Biochem Biophys. 1995 Sep 10;322(1):43-52. PMID:7574693 doi:http://dx.doi.org/10.1006/abbi.1995.1434
- ↑ Plancarte A, Nava G, Mendoza-Hernandez G. Purification, properties, and kinetic studies of cytoplasmic malate dehydrogenase from Taenia solium cysticerci. Parasitol Res. 2009 Jul;105(1):175-83. Epub 2009 Mar 10. PMID:19277715 doi:10.1007/s00436-009-1380-6
- ↑ Goward CR, Nicholls DJ. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994 Oct;3(10):1883-8. PMID:7849603 doi:http://dx.doi.org/10.1002/pro.5560031027
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Jake Ezell, Joel L. Sussman, Joshua Johnson, Angel Herraez, Jaime Prilusky, David Canner