User:Alexander Rudecki/Sandbox 1
From Proteopedia
| Line 5: | Line 5: | ||
DromeQC is encoded by chromosome 3L, locus 64F4-64F5 in the ''D. melanogaster'' genome<ref name="genecard">DromeQC Gene Card. NCBI. [http://www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=Graphics&list_uids=38663]</ref>. It is transcribed into a 1622 nucleotide transcript, containing 5' (36 nucleotides) and 3' (83 nucleotides) untranslated regions <ref name="genecard"/>. The translated protein contains 340 residues corresponding to a M=38,028 Da<ref>DromeQC. UniProt. [http://www.uniprot.org/uniprot/Q9VRQ9]</ref>. It contains a 27 residue signal sequence, suggesting its involvement in the secretory pathway <ref name="schilling"/>. | DromeQC is encoded by chromosome 3L, locus 64F4-64F5 in the ''D. melanogaster'' genome<ref name="genecard">DromeQC Gene Card. NCBI. [http://www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=Graphics&list_uids=38663]</ref>. It is transcribed into a 1622 nucleotide transcript, containing 5' (36 nucleotides) and 3' (83 nucleotides) untranslated regions <ref name="genecard"/>. The translated protein contains 340 residues corresponding to a M=38,028 Da<ref>DromeQC. UniProt. [http://www.uniprot.org/uniprot/Q9VRQ9]</ref>. It contains a 27 residue signal sequence, suggesting its involvement in the secretory pathway <ref name="schilling"/>. | ||
==Structure== | ==Structure== | ||
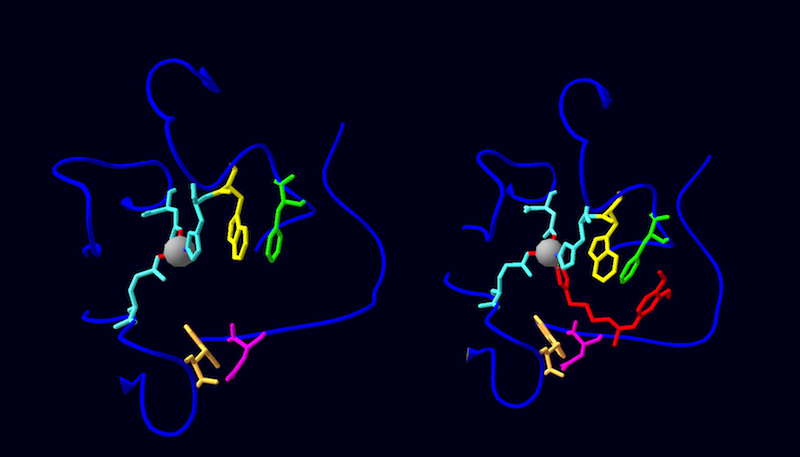
| - | + | DromeQC is made up of 2 identical, independent monomers that come together to form an asymmetric homodimer. The subunits are connected via 4 hydrogen bonds (Chain 1→Chain 2: ARG35 NH2→GLU64 OE2, ARG43 NH2→ASN71 O, ARG43 NH2→PHE75 O, ARG43 NH1→PHE75 O) and surface complementarity. The subunits exhibit a globular α/β-hydrolase fold, characterized by a central twisted β-sheet motif consisting of 5 parallel strands (β1 and β3-β6) and an antiparallel β2 strand. This β-center is flanked by 9 surrounding α-helices; 2 fill the concave face (α5, α7), 7 fill the convex face (α1- α5, α8, α9) with one helix at the edge (α6) of each monomer. Within each subunit is 1 cysteine bond (C113→C136) linking the β3 strand to the α3 helix. These cysteine residues are situated close to the active site, and are conserved in the human orthologue suggesting a pivotal role in catalysis<ref name="schilling"/>. However, when cysteines were replaced with alanines via site-directed mutagenesis, no kinetic differences were observed<ref name="main"/>. In contrast, this mutation did affect structural differences as determined by thermal unfolding experiments<ref name="main"/>. These results correspond to the structural stabilization of this disulfide bond in hQC, and lack of its effect on kinetic activity<ref>Ruiz-Carrillo, D., Koch, B., Parthier, C., Wermann, M., Dambe, T., Buchholz, M., Ludwig, H., Heiser, U., Rahfeld, J., Stubbs, M. T., Schilling, S., and H. Demuth. (2011) Structures of glycosylated mammalian glutaminyl cyclases reveal conformational variability near the active center. Biochemistry. 50: 6280-6288. [http://dx.doi.org/10.1021/bi200249h DOI: 10.1021/bi200249h]</ref>.Each DromeQC monomer chelates a catalytic zinc ion via D131 OD2, E171 OE2, and H297 NE2, and is glycosylated (with up to 7 carbohydrate moieties) at the N42 position. | |
[[Image:Overall structure.jpg|thumb|800px|Figure 1. A 3D graphical representation displaying the homodimer glutaminyl cyclase from ''Drosophila melanogaster'' (PDB: 4F9U). Secondary structure is depicted by red (α-helix) and yellow (β-strand) ribbons, glycosyl groups are coloured pinks, while hydrogen bonds between the two monomers are shown by dotted green lines. The active site of QC contains a chelated zinc ion represented by a gray sphere. Also bound to the active site of this crystal structure and depicted as blue is the inhibitor 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea.]] | [[Image:Overall structure.jpg|thumb|800px|Figure 1. A 3D graphical representation displaying the homodimer glutaminyl cyclase from ''Drosophila melanogaster'' (PDB: 4F9U). Secondary structure is depicted by red (α-helix) and yellow (β-strand) ribbons, glycosyl groups are coloured pinks, while hydrogen bonds between the two monomers are shown by dotted green lines. The active site of QC contains a chelated zinc ion represented by a gray sphere. Also bound to the active site of this crystal structure and depicted as blue is the inhibitor 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea.]] | ||
[[Image:DromeQCActiveSite.png]] | [[Image:DromeQCActiveSite.png]] | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 20:27, 30 March 2014
|
Contents |
Introduction
Drosophila melanogaster glutaminyl cyclase (DromeQC but also known as CG10487 CG32412 or Dmel\CG32412) is a globular protein part of the α/β-hydrolase superfamily. DromeQC is an aminoacyltransferase (EC 2.3.2.5) that acts on N-terminal glutamine or glutamate residues, producing a stable cap resistant to protease degradation. The human orthologue of DromeQC (hQC) has been implicated in stabilizing amyloid Aβ peptides involved in neurodegenerative disorders such as Alzheimers[1]. It has been shown that DromeQC has a similar overall fold to hQC, as well as a conserved active site[2]. Thus DromeQC is an attractive candidate for transgenic models and mechanistic studies.
Gene->Protein
DromeQC is encoded by chromosome 3L, locus 64F4-64F5 in the D. melanogaster genome[3]. It is transcribed into a 1622 nucleotide transcript, containing 5' (36 nucleotides) and 3' (83 nucleotides) untranslated regions [3]. The translated protein contains 340 residues corresponding to a M=38,028 Da[4]. It contains a 27 residue signal sequence, suggesting its involvement in the secretory pathway [1].
Structure
DromeQC is made up of 2 identical, independent monomers that come together to form an asymmetric homodimer. The subunits are connected via 4 hydrogen bonds (Chain 1→Chain 2: ARG35 NH2→GLU64 OE2, ARG43 NH2→ASN71 O, ARG43 NH2→PHE75 O, ARG43 NH1→PHE75 O) and surface complementarity. The subunits exhibit a globular α/β-hydrolase fold, characterized by a central twisted β-sheet motif consisting of 5 parallel strands (β1 and β3-β6) and an antiparallel β2 strand. This β-center is flanked by 9 surrounding α-helices; 2 fill the concave face (α5, α7), 7 fill the convex face (α1- α5, α8, α9) with one helix at the edge (α6) of each monomer. Within each subunit is 1 cysteine bond (C113→C136) linking the β3 strand to the α3 helix. These cysteine residues are situated close to the active site, and are conserved in the human orthologue suggesting a pivotal role in catalysis[1]. However, when cysteines were replaced with alanines via site-directed mutagenesis, no kinetic differences were observed[2]. In contrast, this mutation did affect structural differences as determined by thermal unfolding experiments[2]. These results correspond to the structural stabilization of this disulfide bond in hQC, and lack of its effect on kinetic activity[5].Each DromeQC monomer chelates a catalytic zinc ion via D131 OD2, E171 OE2, and H297 NE2, and is glycosylated (with up to 7 carbohydrate moieties) at the N42 position.
References
- ↑ 1.0 1.1 1.2 Schilling S, Zeitschel U, Hoffmann T, Heiser U, Francke M, Kehlen A, Holzer M, Hutter-Paier B, Prokesch M, Windisch M, Jagla W, Schlenzig D, Lindner C, Rudolph T, Reuter G, Cynis H, Montag D, Demuth HU, Rossner S. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer's disease-like pathology. Nat Med. 2008 Oct;14(10):1106-11. doi: 10.1038/nm.1872. Epub 2008 Sep 28. PMID:18836460 doi:http://dx.doi.org/10.1038/nm.1872
- ↑ 2.0 2.1 2.2 Koch B, Kolenko P, Buchholz M, Ruiz Carrillo D, Parthier C, Wermann M, Rahfeld JU, Reuter G, Schilling S, Stubbs MT, Demuth HU. Crystal Structures of Glutaminyl Cyclases from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs. Biochemistry. 2012 Aug 16. PMID:22897232 doi:10.1021/bi300687g
- ↑ 3.0 3.1 DromeQC Gene Card. NCBI. [1]
- ↑ DromeQC. UniProt. [2]
- ↑ Ruiz-Carrillo, D., Koch, B., Parthier, C., Wermann, M., Dambe, T., Buchholz, M., Ludwig, H., Heiser, U., Rahfeld, J., Stubbs, M. T., Schilling, S., and H. Demuth. (2011) Structures of glycosylated mammalian glutaminyl cyclases reveal conformational variability near the active center. Biochemistry. 50: 6280-6288. DOI: 10.1021/bi200249h