We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Daud Akhtar/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
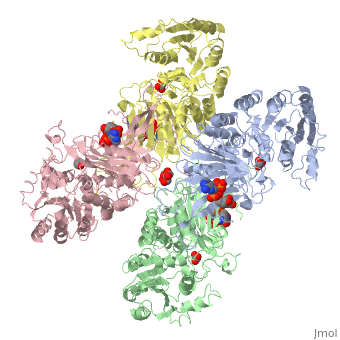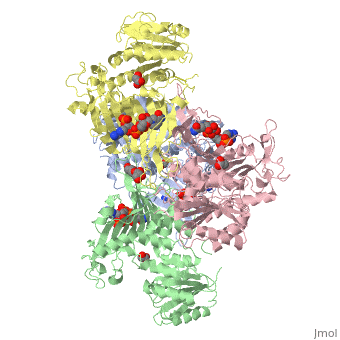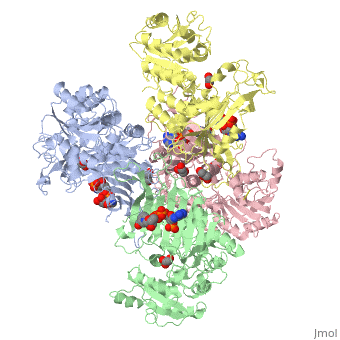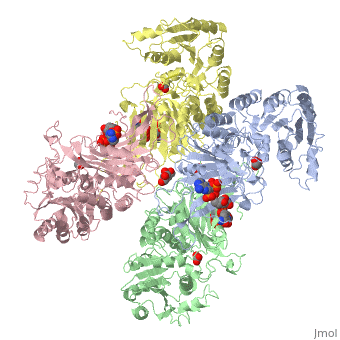
G6PD belongs to the Glucose-6-phosphate dehydrogenase-like family of proteins, which are characterized by rossmann-like domains. It also belongs to the superfamily GAPDH-like domain, which consists of 2-domain proteins with an alpha+beta domain. The overall structure of human G6PD is present as a homodimer/homotetramer equilibrium that is dependant on pH and ionic strength. The individual monomers appear to be inactive where each monomer consists of 514 amino acids with a molecular weight of 59kDa. At high pH and ionic strength, the equilibrium is shifted towards the dimer, whereas low pH conditions cause a shift to the tetramer<ref name="1qki" />. Specifically, G6PD has a structural NADP+ moiety next to a separate catalytic site for NADP+. Cohen (1968) showed that NADP+ binding stabilizes the hydrophobic interactions between subunits, thus preventing disassociation of the dimer state into monomers. Crystallization experiments by Au (2000) using the Canton Arg459->Leu (R459L) which is the most common Chinese variant, showed the Canton R459L G6PD enzyme as a dimer of dimers where each specific monomer consists of two domains<ref name="1qki" />. | G6PD belongs to the Glucose-6-phosphate dehydrogenase-like family of proteins, which are characterized by rossmann-like domains. It also belongs to the superfamily GAPDH-like domain, which consists of 2-domain proteins with an alpha+beta domain. The overall structure of human G6PD is present as a homodimer/homotetramer equilibrium that is dependant on pH and ionic strength. The individual monomers appear to be inactive where each monomer consists of 514 amino acids with a molecular weight of 59kDa. At high pH and ionic strength, the equilibrium is shifted towards the dimer, whereas low pH conditions cause a shift to the tetramer<ref name="1qki" />. Specifically, G6PD has a structural NADP+ moiety next to a separate catalytic site for NADP+. Cohen (1968) showed that NADP+ binding stabilizes the hydrophobic interactions between subunits, thus preventing disassociation of the dimer state into monomers. Crystallization experiments by Au (2000) using the Canton Arg459->Leu (R459L) which is the most common Chinese variant, showed the Canton R459L G6PD enzyme as a dimer of dimers where each specific monomer consists of two domains<ref name="1qki" />. | ||
| - | [[Image: | + | [[Image:Tetramer_without_title.png|thumb|500px|Figure 1: Cartoon figure of the first biological assemblye unit of glucose-6-phosphate dehydrogenase(PDBID:1QKI) indicating 4 identical monomers which are coloured by chain(A=yellow, B=blue, C=green and D=red. SWISSPDBViewer was used to generate the above image.]] |
| Line 22: | Line 22: | ||
===Substrate Binding and Catalytic Mechanism=== | ===Substrate Binding and Catalytic Mechanism=== | ||
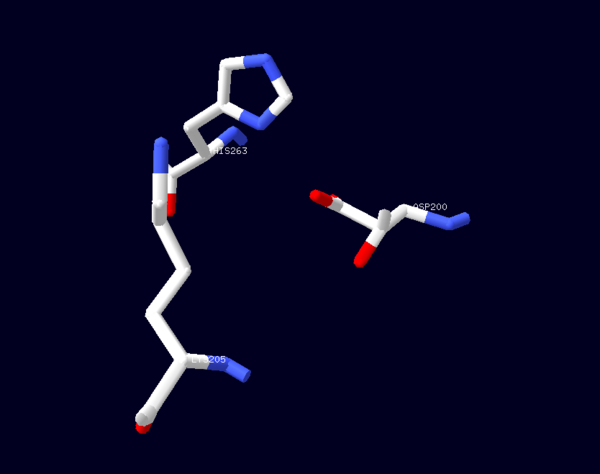
The catalytic mechanism involved with G6PD is a general acid/base reaction. It begins with G6P binding to its active site in the second domain by interacting with Lys205. His 263 then acts as a general base that abstracts the proton from the C1-hydroxyl of G6P. Cosgrove (1998) suggest that Asp200 stabilizes the positive charge on the N atom of His264 in the transition state forming a catalytic dyad. Asp200 also plays a role in binding the phosphate moiety of G6P<ref>PMID: 9485426 </ref>. | The catalytic mechanism involved with G6PD is a general acid/base reaction. It begins with G6P binding to its active site in the second domain by interacting with Lys205. His 263 then acts as a general base that abstracts the proton from the C1-hydroxyl of G6P. Cosgrove (1998) suggest that Asp200 stabilizes the positive charge on the N atom of His264 in the transition state forming a catalytic dyad. Asp200 also plays a role in binding the phosphate moiety of G6P<ref>PMID: 9485426 </ref>. | ||
| - | [[Image: | + | [[Image:Cat dyad without.png|thumb|600px|Figure 2: A stick representation of the amino acid residues (Lys205, His263,Asp200) involved in the catalysis reaction associated with Glucose-6-Phosphate Dehydrogenase (PDB ID: 1QKI): CPK notation was used to depict the catalytic dyad. Image was generated using SWISSPDBViewer |
]] | ]] | ||
==Biological Role== | ==Biological Role== | ||
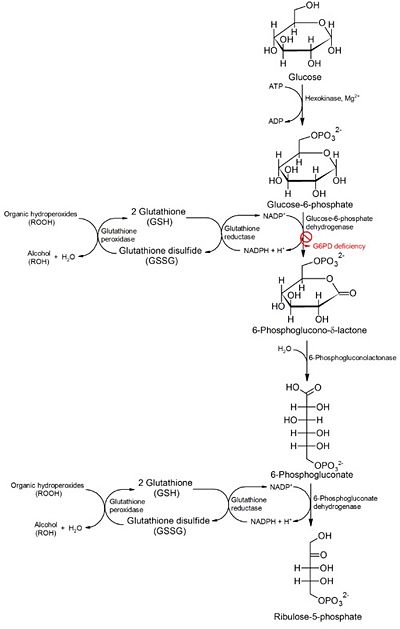
The major catabolic fate of G6P is glycolytic breakdown through glycolysis. Another fate however is the oxidation of G6P into pentose phosphates through the pentose phosphate pathway. The first reaction of the pentose phosphate pathway is the oxidation of G6P by G6PD to form 6-phosphoglucono-δ-lactone. NADP+ is the electron acceptor, and the overall equilibrium for this reaction favours NADPH formation. In this pathway, G6PD is the rate-limiting enzyme. Biologically, the pentose phosphate pathway plays an important role in generating NADPH. High levels of NADP+ in the cytosol act as an allosteric stimulator of G6PD by promoting G6P usage as a substrate in the pentose phosphate pathway. The ultimate product of the pentose phosphate pathway is ribose-5- phosphate. Rapidly dividing cells, such as those of bone marrow, skin, and tumors use ribose-5-phosphate to make RNA, DNA, and coenzymes such as ATP, NADH, and FADH2<ref>PMID: 12838507 </ref>. In other tissues, the essential product of the pentose phosphate pathway is not the pentose products but the electron donors NADPH. NADPH is needed for reductive biosynthesis or to counter the damaging effects of oxygen radicals. Additionally tissues that are heavily involved in fatty acid synthesis such as liver and adipose tissue, require NADPH to fuel the synthesis of cholesterols and steroid hormones<ref>PMID: 9553122 </ref>. In the case of erythrocytes and cells which are directly exposed to oxygen, a high ratio of NADPH/NADP+ and glutathione prevents or undoes oxidative damage caused by the generation of free radicals. A genetic defect in G6PD is known as G6PD Deficiency and can have serious medical consequences <ref name="1qki" /><ref>PMID: 2018843</ref> | The major catabolic fate of G6P is glycolytic breakdown through glycolysis. Another fate however is the oxidation of G6P into pentose phosphates through the pentose phosphate pathway. The first reaction of the pentose phosphate pathway is the oxidation of G6P by G6PD to form 6-phosphoglucono-δ-lactone. NADP+ is the electron acceptor, and the overall equilibrium for this reaction favours NADPH formation. In this pathway, G6PD is the rate-limiting enzyme. Biologically, the pentose phosphate pathway plays an important role in generating NADPH. High levels of NADP+ in the cytosol act as an allosteric stimulator of G6PD by promoting G6P usage as a substrate in the pentose phosphate pathway. The ultimate product of the pentose phosphate pathway is ribose-5- phosphate. Rapidly dividing cells, such as those of bone marrow, skin, and tumors use ribose-5-phosphate to make RNA, DNA, and coenzymes such as ATP, NADH, and FADH2<ref>PMID: 12838507 </ref>. In other tissues, the essential product of the pentose phosphate pathway is not the pentose products but the electron donors NADPH. NADPH is needed for reductive biosynthesis or to counter the damaging effects of oxygen radicals. Additionally tissues that are heavily involved in fatty acid synthesis such as liver and adipose tissue, require NADPH to fuel the synthesis of cholesterols and steroid hormones<ref>PMID: 9553122 </ref>. In the case of erythrocytes and cells which are directly exposed to oxygen, a high ratio of NADPH/NADP+ and glutathione prevents or undoes oxidative damage caused by the generation of free radicals. A genetic defect in G6PD is known as G6PD Deficiency and can have serious medical consequences <ref name="1qki" /><ref>PMID: 2018843</ref> | ||
| - | [[Image: | + | [[Image:Pathology of G6PD deficiency.jpg|thumb|400px|left|Figure 3: A diagram of the pentose phosphate pathway showing the conversion of glucose into ribose-5-phosphate (source http://en.wikipedia.org/wiki/File:Pathology_of_G6PD_deficiency.png)]] |
Revision as of 01:34, 1 April 2014
Glucose-6-Phosphate Dehydrogenase(G6PD)
| |||||||||||
References
- ↑ Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr. 2001;21:121-40. PMID:11375432 doi:http://dx.doi.org/10.1146/annurev.nutr.21.1.121
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Au SW, Gover S, Lam VM, Adams MJ. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure. 2000 Mar 15;8(3):293-303. PMID:10745013
- ↑ Kotaka M, Gover S, Vandeputte-Rutten L, Au SW, Lam VM, Adams MJ. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005 May;61(Pt 5):495-504. Epub 2005, Apr 20. PMID:15858258 doi:http://dx.doi.org/10.1107/S0907444905002350
- ↑ Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupianez JA, Del Rio LA. A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J. 1998 Mar 1;330 ( Pt 2):777-84. PMID:9480890
- ↑ Au SW, Naylor CE, Gover S, Vandeputte-Rutten L, Scopes DA, Mason PJ, Luzzatto L, Lam VM, Adams MJ. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr D Biol Crystallogr. 1999 Apr;55(Pt 4):826-34. PMID:10089300
- ↑ Bhadbhade MM, Adams MJ, Flynn TG, Levy HR. Sequence identity between a lysine-containing peptide from Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase and an active site peptide from human erythrocyte glucose-6-phosphate dehydrogenase. FEBS Lett. 1987 Jan 26;211(2):243-6. PMID:3100332
- ↑ Cosgrove MS, Naylor C, Paludan S, Adams MJ, Levy HR. On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase. Biochemistry. 1998 Mar 3;37(9):2759-67. PMID:9485426 doi:10.1021/bi972069y
- ↑ Ramos KL, Colquhoun A. Protective role of glucose-6-phosphate dehydrogenase activity in the metabolic response of C6 rat glioma cells to polyunsaturated fatty acid exposure. Glia. 2003 Aug;43(2):149-66. PMID:12838507 doi:http://dx.doi.org/10.1002/glia.10246
- ↑ Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998 Apr 24;273(17):10609-17. PMID:9553122
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991 May 1;77(9):2059-64. PMID:2018843
- ↑ . Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ. 1989;67(6):601-11. PMID:2633878
- ↑ Manganelli G, Masullo U, Passarelli S, Filosa S. Glucose-6-phosphate dehydrogenase deficiency: disadvantages and possible benefits. Cardiovasc Hematol Disord Drug Targets. 2013 Mar 1;13(1):73-82. PMID:23534950
- ↑ Beutler E. Glucose-6-phosphate dehydrogenase deficiency. N Engl J Med. 1991 Jan 17;324(3):169-74. PMID:1984194 doi:http://dx.doi.org/10.1056/NEJM199101173240306