We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox reserved 915
From Proteopedia
(Difference between revisions)
| Line 35: | Line 35: | ||
====Binding==== | ====Binding==== | ||
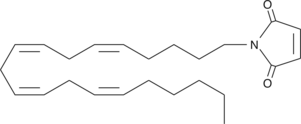
| - | 2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive. '''See Overall Reaction.''' | + | 2-AG binds to the catalytic triad and is hydrolyzed. The structure of 2-AG contains a long and flexible aliphatic chain and a polar head that is cleaved. It is the polar head that gets attracted to the catalytic triad and binds to it so the catalytic Serine can cleave 2-AG. 2-AG is broken down into arachidonic acid and glycerol which makes 2-AG inactive<ref name="Bertrand" />. '''See Overall Reaction.''' |
===Inhibition of Catalytic Triad=== | ===Inhibition of Catalytic Triad=== | ||
Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This is a natural inhibition phenomenon. Inhibition of MGL leads to increase in 2-AG levels since AG is broken down by MGL <ref name="Clemente" />. The catalytic triad is a major part of MGL and its interaction with other parts within the brain and how the brain functions. | Research on MGL is being geared towards inhibiting 2-AG from binding to the catalytic triad and being hydrolyzed. The binding of 2-AG to the catalytic triad can be extracted before being hydrolyzed. MPD (2-methyl-pentane-2,4-diol)is located at the end of the tunnel where the catalytic triad is at and the tunnel is filled with MPD molecules. MPD being in the same vicinity will extract 2-AG from the triad and the MPD molecule will sit in there in place of 2-AG. This is a natural inhibition phenomenon. Inhibition of MGL leads to increase in 2-AG levels since AG is broken down by MGL <ref name="Clemente" />. The catalytic triad is a major part of MGL and its interaction with other parts within the brain and how the brain functions. | ||
| + | |||
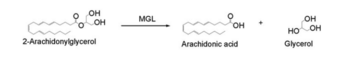
| + | ===Overall Reaction=== | ||
| + | [[Image:Reaction.PNG|350px|thumb|Overall reaction representing the hydrolysis of 2-AG by MGL]] | ||
| + | In this reaction 2-AG binds to the catalytic triad in the oxyanion hole in the active site. In the [[:Category:Oxyanion hole| oxyanion holes]] the substrate is stabilized by two nitrogen atoms from the catalytic Hisdidine and Asperatate during the transition step of the catalytic reaction. The catalytuc triad activates the nucleophilic serine and cleaves the ester bond of 2-AG that is being stabilized by its carbonyl group that is attached to the oxyanion hole. The glycerol molecule is released and it might diffuse to the narrow "exit hole", while the arachidonic acid would diffuse back to the top of the tunnel and leave the protein <ref name="Bertrand" />. | ||
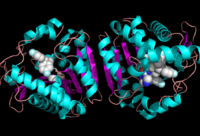
==Ligand Binding Site== | ==Ligand Binding Site== | ||
[[Image:Overall_ligand.png|left|200px|thumb|Ligand within the Overall Structure of MGL]] | [[Image:Overall_ligand.png|left|200px|thumb|Ligand within the Overall Structure of MGL]] | ||
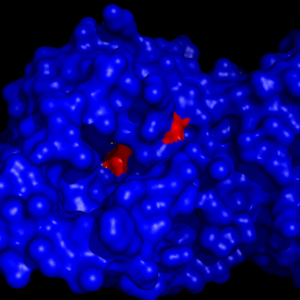
The <scene name='58/580298/Ligand/1'>ligand binding pocket</scene> of MGL has a large hydrophobic region with a polar bottom. The entrance of the binding pocket for MGL contains a lid, which is very flexible. The binding pocket or tunnel within MGL matches with the overall structure of 2-AG, with 2-AG's polar head being cleaved by the catalytic triad. Bertrand found that in MGL the binding pocket is not adjusted to the ligand's shape. However, the main movements of MGL associated with ligand binding involved the lid region. When 2-AG and its isomer 1(3)-AG bind to MGL, the hydrophobic chain is first aligned with the left part of the binding pocket. The carbonyl is then hydrogen bonded to <scene name='58/580298/Ala61/1'>Ala61</scene>. The polar head group of the ligand is then fixed by three hydrogen bonds. As a result, future research is looking into the large lipophilic portion of the binding pocket for designing selective inhibitors <ref name="Bertrand" />. | The <scene name='58/580298/Ligand/1'>ligand binding pocket</scene> of MGL has a large hydrophobic region with a polar bottom. The entrance of the binding pocket for MGL contains a lid, which is very flexible. The binding pocket or tunnel within MGL matches with the overall structure of 2-AG, with 2-AG's polar head being cleaved by the catalytic triad. Bertrand found that in MGL the binding pocket is not adjusted to the ligand's shape. However, the main movements of MGL associated with ligand binding involved the lid region. When 2-AG and its isomer 1(3)-AG bind to MGL, the hydrophobic chain is first aligned with the left part of the binding pocket. The carbonyl is then hydrogen bonded to <scene name='58/580298/Ala61/1'>Ala61</scene>. The polar head group of the ligand is then fixed by three hydrogen bonds. As a result, future research is looking into the large lipophilic portion of the binding pocket for designing selective inhibitors <ref name="Bertrand" />. | ||
| - | |||
| - | ==Overall Reaction== | ||
| - | [[Image:Reaction.PNG|350px|thumb|Overall reaction representing the hydrolysis of 2-AG by MGL]] | ||
| - | In this reaction 2-AG binds to the catalytic triad in the oxyanion hole in the active site. In the [[:Category:Oxyanion hole| oxyanion holes]] the oxygen of the substrate is stabilized by two nitrogen atoms during the transition step of the catalytic reaction. The triad activates the nucleophilic serine and cleaves the ester bond of 2-AG that is being stabilized by its carbonyl group that is attached to the oxyanion hole. The glycerol molecule is released and it might diffuse to the narrow "exit hole", while the arachidonic acid would diffuse back to the top of the tunnel and leave the protein <ref name="Bertrand" />. | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 21:54, 8 April 2014
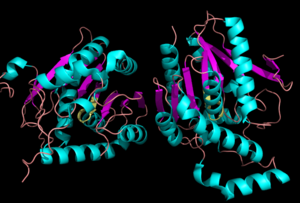
Monoglyceride Lipase (MGL)
| |||||||||||