Porin
From Proteopedia
| Line 1: | Line 1: | ||
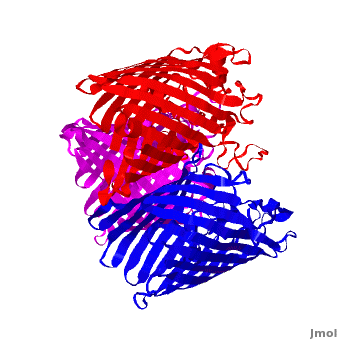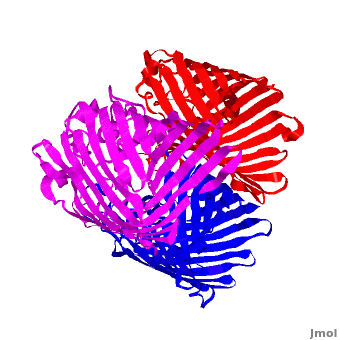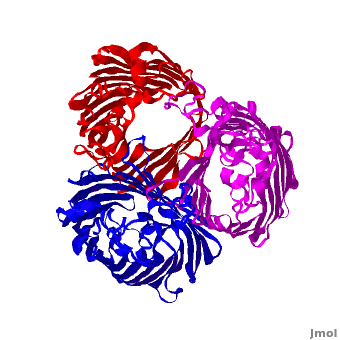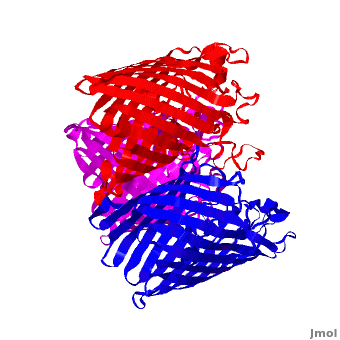
<StructureSection load='2j1n' size='450' side='right' scene='Porin/Cv/1' caption='E. coli Ompc (PDB code [[2j1n]])'> | <StructureSection load='2j1n' size='450' side='right' scene='Porin/Cv/1' caption='E. coli Ompc (PDB code [[2j1n]])'> | ||
| - | + | ||
{{Clear}} | {{Clear}} | ||
[[Porin]] or '''Outer Membrane Proteins''' '''(Omps)''' act as channels which allow passive diffusion of sugars, ions and amino acids. They are beta barrel proteins which traverse the cell membrane. In ''E. coli'' they are named according to their genes: C, F, G (OmpC, OmpF, Ompg). See more details in<br /> | [[Porin]] or '''Outer Membrane Proteins''' '''(Omps)''' act as channels which allow passive diffusion of sugars, ions and amino acids. They are beta barrel proteins which traverse the cell membrane. In ''E. coli'' they are named according to their genes: C, F, G (OmpC, OmpF, Ompg). See more details in<br /> | ||
| Line 10: | Line 10: | ||
<scene name='1a0s/Hidrophobic/1'>seen</scene> from the hydrophobic ring around the protein, this makes it possible to submerge in the lipid bilayer (hydrophobic amino acids are sandybrown, hydrophilic ones are cyan). As you can <scene name='1a0s/Hidrophobic1/1'>see</scene> the hole in the protein is made of mainly hydrophilic chains thus making it possible for the sugar to pass through (these scenes were created by Nádori Gergely). | <scene name='1a0s/Hidrophobic/1'>seen</scene> from the hydrophobic ring around the protein, this makes it possible to submerge in the lipid bilayer (hydrophobic amino acids are sandybrown, hydrophilic ones are cyan). As you can <scene name='1a0s/Hidrophobic1/1'>see</scene> the hole in the protein is made of mainly hydrophilic chains thus making it possible for the sugar to pass through (these scenes were created by Nádori Gergely). | ||
| - | __NOTOC__ | ||
=== Gating and conduction of <scene name='Journal:JBSD:3/Cv/7'>nano-channel forming proteins</scene>, a computational approach <ref>doi 10.1080/07391102.2012.712460</ref>=== | === Gating and conduction of <scene name='Journal:JBSD:3/Cv/7'>nano-channel forming proteins</scene>, a computational approach <ref>doi 10.1080/07391102.2012.712460</ref>=== | ||
The functional units of the living systems are cells whose internal physico-chemical conditions needed for optimum function are different from that of the external medium and are maintained by hydrophobic membrane barrier and reconstituted water filled nano-pore forming proteins. The structure of these channels dictates their function to some extent and makes them to open or close in response to various conditions in the surrounding medium including pH, temperature, ionic strength, potential difference, osmotic pressure, presence of certain ligands and so on. Due to very complex and sensitive structures of these molecules to the medium and the effect of their native location, lipid Bilayer, different from soluble proteins, the molecular structure of most of membrane proteins have not been worked out at atomic level yet. The discovery of the crystal structure of certain membrane macromolecules have paved the way to understand the mechanism(s) by which they control the traffic of certain molecules through the membrane and the way they respond to the internal and external stimulus and signal transduction. | The functional units of the living systems are cells whose internal physico-chemical conditions needed for optimum function are different from that of the external medium and are maintained by hydrophobic membrane barrier and reconstituted water filled nano-pore forming proteins. The structure of these channels dictates their function to some extent and makes them to open or close in response to various conditions in the surrounding medium including pH, temperature, ionic strength, potential difference, osmotic pressure, presence of certain ligands and so on. Due to very complex and sensitive structures of these molecules to the medium and the effect of their native location, lipid Bilayer, different from soluble proteins, the molecular structure of most of membrane proteins have not been worked out at atomic level yet. The discovery of the crystal structure of certain membrane macromolecules have paved the way to understand the mechanism(s) by which they control the traffic of certain molecules through the membrane and the way they respond to the internal and external stimulus and signal transduction. | ||
Revision as of 07:06, 20 August 2014
| |||||||||||
3D structures of Porin
Updated on 20-August-2014
2xe5, 2xe1, 2xe2, 2xe3, 2j1n – EcOmpC – Escherichia coli
3k19, 3k1b, 2zfg, 1omf, 2omf, 1opf, 3poq, 3pou, 3pox, 4jfb, 4lse, 4lsf, 4lsh, 4lsi – EcOmpF
3nb3 – EcOmpA + EcOmpC - EM
3upg, 3uu2 – OmpC – Salmonella enterica
1hxt, 1hxu, 1hxx, 1bt9, 1gfm, 1gfn, 1gfo, 1gfp, 1gfq, 1mpf – EcOmpF (mutant)
2iwv, 2iww, 2f1c - EcOmpG
2x9k - EcOmpG (mutant)
2wjq, 2wjr – EcOmp NANC
1pho - EcOmp
3a2r – NmOmpB – Neisseria meningitides
3sy9 – PaOpdC – Pseudomonas aeruginosa
2odj, 3sy7, 4foz – PaOprD
3szd - PaOpdF
3t20 – PaOpdH
2qtk, 2y2x, 3sys – PaOpdK
3t0s - PaOpdL
2y0k, 2y0l, 3szv – PaOpdO
3syb – PaOpdP
3t24 - PaOpdQ
3jty – Omp – Pseudomonas fluorescens
2v9u – MsOmp rim domain – Mycobacteria smegmatis
1uun – MsOmp (mutant)
2fgr - DaOmp32 - Delftia acidovorans
1a0s, 1mpr - StOmp - Salmonella typhimurium
3nsg - StOmpF
2prn, 1bh3, 3prn, 5prn, 6prn, 7prn, 8prn – RbOmp (mutant) - Rhodopseudomonas blastica
1prn - RbOmp
1osm – OmpK36 – Klebsiella pneumoniae
2por, 3por – Omp – Rhodobacter capsulatus
4aui – Omp – Neisseria gonorrheae
4gey, 4gf4 – Omp B – Pseudomonas putida
Porin + polypeptides
2j4u - EcOmpC + lactotransferrin fragment
2zld - EcOmpF + colicin E3
3o0e - EcOmpF + colicin peptide
1h6s - RbOmp + inserted sequence
Porin + various compounds
3hw9, 3hwb - EcOmpF + ions
3fyx - Ec~OmpF + dibenzo-18-crown-6
4gcp, 4gcq, 4gcs – EcOmpF + antibiotic
1af6, 1mpq, 1mpm, 1mpn, 1mpo - EcOmp LAMB + sugars
3a2s, 3a2t - NmOmpB + sugars
3a2u - NmOmpB + AMPPNP
2o4v - PaOprP + phosphate
2fgq - DaOmp32 + malate
1e54 - Omp32 + sulfate - Comamonas acidovorans
1oh2, 1a0t - StOmp + sucrose
Voltage-Dependent Anion Channel
2jk4 – hVDAC-1 – human
2k4t - hVDAC-1 - NMR
3emn – VDAC-1 - mouse
References
- ↑ Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006 Oct 6;362(5):933-42. Epub 2006 Aug 3. PMID:16949612 doi:10.1016/j.jmb.2006.08.002
- ↑ Besya AB, Mobasheri H, Ejtehadi MR. Gating and conduction of nano-channel forming proteins: a computational approach. J Biomol Struct Dyn. 2012 Aug 28. PMID:22928968 doi:10.1080/07391102.2012.712460
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky