Sandbox Reserved 911
From Proteopedia
| Line 14: | Line 14: | ||
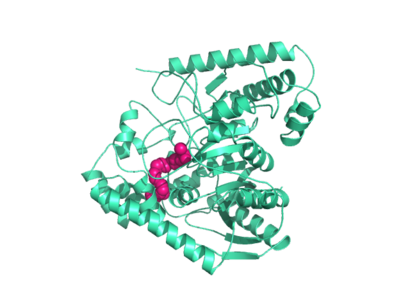
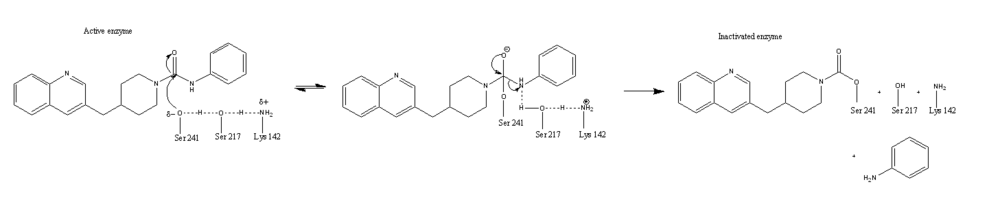
Mutagenesis and inhibitor studies have shown that FAAH has a <scene name='57/573125/2vya/15'>Ser-Ser-Lys catalytic triad</scene>, consisting of Ser241, Ser217, and Lys142 <ref name="1MT5"/>. Ser-Ser-Lys catalytic triads are not often seen in hydrolases, making FAAH an enzyme of interest for additional research to better determine how proteins with this catalytic triad function. Ser241 acts as the catalytic nucleophile for the cleavage of amide bonds (Figure 2) <ref name="1MT5"/>. [http://proteopedia.org/wiki/index.php/Fatty_acid_amide_hydrolase Inhibitors] are able to inactivate the catalytic triad by providing a substrate containing a leaving group, such as aniline, that is a more favorable leaving group than the Ser241 hydroxyl group. With the serine bound to the carbonyl carbon, FAAH is no longer able to accommodate any more substrates <ref name="2VYA"/>. FAAH also requires two water molecules in its active site to properly position and cleave amide bonds. One water molecule (W1) deacylates the substrate, and the other (W2) helps coordinate W1 through the catalytic K142 (Figure 3) <ref name="3LJ6">PMID:20493882</ref> . | Mutagenesis and inhibitor studies have shown that FAAH has a <scene name='57/573125/2vya/15'>Ser-Ser-Lys catalytic triad</scene>, consisting of Ser241, Ser217, and Lys142 <ref name="1MT5"/>. Ser-Ser-Lys catalytic triads are not often seen in hydrolases, making FAAH an enzyme of interest for additional research to better determine how proteins with this catalytic triad function. Ser241 acts as the catalytic nucleophile for the cleavage of amide bonds (Figure 2) <ref name="1MT5"/>. [http://proteopedia.org/wiki/index.php/Fatty_acid_amide_hydrolase Inhibitors] are able to inactivate the catalytic triad by providing a substrate containing a leaving group, such as aniline, that is a more favorable leaving group than the Ser241 hydroxyl group. With the serine bound to the carbonyl carbon, FAAH is no longer able to accommodate any more substrates <ref name="2VYA"/>. FAAH also requires two water molecules in its active site to properly position and cleave amide bonds. One water molecule (W1) deacylates the substrate, and the other (W2) helps coordinate W1 through the catalytic K142 (Figure 3) <ref name="3LJ6">PMID:20493882</ref> . | ||
| - | [[Image:Catalytic_triad2.png|1000px|left|thumb|Figure 2: Catalytic Triad and PF-750 Inhibitor Mechanism; The combination of Ser241, Ser217, and | + | [[Image:Catalytic_triad2.png|1000px|left|thumb|Figure 2: Catalytic Triad and PF-750 Inhibitor Mechanism; The combination of Ser241, Ser217, and Lys142 provides a partial negative charge on the Ser241 hydroxyl group. The Ser241 oxygen attacks the carbonyl carbon of PF-750, resulting in a tetrahedral intermediate. Aniline is released as a leaving group, with the remaining portion of PF-750 still covalently bound to Ser241. FAAH is now inactivated, unable to accommodate any new ligands <ref name="2VYA"/>. ]] |
==Relationship to other proteins== | ==Relationship to other proteins== | ||
Revision as of 03:15, 17 April 2014
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
| |||||||||||
Applications
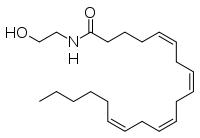
The human nervous system has several types of chemical messengers, including amino acids, lipids, peptide hormones, and monoamines [1]. FAAH primarily degrades anandamide (AEA), a naturally-occurring signaling lipid that functions in the brain. AEA brings pain relief to the body. Inhibiting FAAH would likely sustain AEA signaling, leading to prolonged pain relief and decreased inflammation [2].
FAAH plays a role in endocannabinoid signaling that has intriguing potential as a drug target. This signaling system consists of endocannabinoid ligands (such as AEA), two G protein-coupled receptors (CB1 and CB2), and the enzymes that synthesize and degrade (such as FAAH) the signaling lipids. Previous research has explored the potential of regulating endocannabinoid signaling through the CB1 and CB2 receptors. However, molecules found to activate these receptors (such as tetrahydrocannabinol (THC), the main psychoactive ingredient of marijuana), while providing the intended pain relief, also produce many undesirable side effects, such as decreased cognition and motor control. On the other hand, research involving FAAH inhibitors has shown that blocking this part of the pathway reduces pain without the unwanted side effects seen through CB1/CB2 activation. Thus, exploring the possibility of using FAAH inhibition to decrease pain relief with minimal side effects could lead to new pain treatment solutions [2].
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002 Nov 29;298(5599):1793-6. PMID:12459591 doi:10.1126/science.1076535
- ↑ 2.0 2.1 2.2 2.3 Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009 Apr 24;16(4):411-20. PMID:19389627 doi:10.1016/j.chembiol.2009.02.013
- ↑ 3.0 3.1 3.2 3.3 Mileni M, Johnson DS, Wang Z, Everdeen DS, Liimatta M, Pabst B, Bhattacharya K, Nugent RA, Kamtekar S, Cravatt BF, Ahn K, Stevens RC. Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc Natl Acad Sci U S A. 2008 Sep 2;105(35):12820-4. Epub 2008 Aug 27. PMID:18753625
- ↑ Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Binding and inactivation mechanism of a humanized fatty acid amide hydrolase by alpha-ketoheterocycle inhibitors revealed from cocrystal structures. J Am Chem Soc. 2009 Aug 5;131(30):10497-506. PMID:19722626 doi:10.1021/ja902694n
- ↑ 5.0 5.1 5.2 Mileni M, Kamtekar S, Wood DC, Benson TE, Cravatt BF, Stevens RC. Crystal structure of fatty acid amide hydrolase bound to the carbamate inhibitor URB597: discovery of a deacylating water molecule and insight into enzyme inactivation. J Mol Biol. 2010 Jul 23;400(4):743-54. Epub 2010 May 21. PMID:20493882 doi:10.1016/j.jmb.2010.05.034