We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Brittany Carroll/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
== Classification == | == Classification == | ||
| - | + | CATH (link out) | |
1. | 1. | ||
2. | 2. | ||
| Line 27: | Line 27: | ||
== Structural highlights == | == Structural highlights == | ||
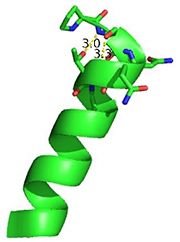
| + | N-terminal helix cap | ||
| + | [[Image:Ntermcap.jpg|300|left|thumb]] | ||
| + | The N-terminal cap of the helix follows a Ib motiff. This motiff is also known as a capping box. | ||
| + | |||
| + | N’ -> N4 h-xpxph | ||
| + | N’- P S N Q T L –N4 (residues 135-140 in 3otb) | ||
| + | |||
| + | Hydrogen bonds between N’ and the backbone of N3 and N3 with N’ backbone are shown in the figure. The figure is difficult to see the T with P bb but it is not linear, this may just be due to modeling as it is close enough to form a h-bond. There is also a hydrophobic interaction between P and L. | ||
| + | |||
| + | [[Image:Catpi.jpg|300|Right|thumb|This image depicts two cation-π interactions between Arg and Tyr or Trp. The energetic significances are -1.22 and -6.55 kj/mol respectively. (site website) (30tb)]] | ||
| + | A cation-π interaction occurs between a cation and the face of a simple aromatic, there is partial negative charge in the center of the ring. The cation-π interaction is actually stronger than a salt bridge because of the desolvation penalty. With the cation-π interaction the cation has a similar dosolvation penalty to pay as the salt bridge ions but the π system is already poorly solvated. Also there is not neutralization of charge that occurs between the two groups. These properties of the cation-π interaction imply that thecation-π interactions on protein surfaces (mainly where they are seen) could contribute to protein structure and stability. | ||
| - | == Headline text == | ||
Revision as of 15:59, 24 April 2014
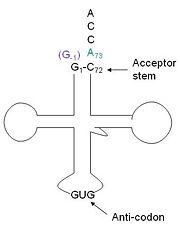
tRNA(His) guanylyltransferase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644