This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Abbas Raza/Sandbox 1
From Proteopedia
| Line 18: | Line 18: | ||
== '''Function''' == | == '''Function''' == | ||
| - | MmuNeil3 is a bifunctional DNA glycosylase that exhibits a broad substrate recognition spectrum (Sp, Gh, FapyG, FapyA, MeFapyG, DHU, DHT, 5-OHU, 5-OHC,5-OHMH, Tg, 8-oxoA, AP) but not 8-oxo-G.[[Image: | + | MmuNeil3 is a bifunctional DNA glycosylase that exhibits a broad substrate recognition spectrum (Sp, Gh, FapyG, FapyA, MeFapyG, DHU, DHT, 5-OHU, 5-OHC,5-OHMH, Tg, 8-oxoA, AP) but not 8-oxo-G. |
| + | [[Image:dna_lesions.jpg]] | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
Revision as of 21:00, 27 April 2014
Contents |
NEIL3: a DNA repair glycosylase from Mus musculus
|
This protein is a unique member of DNA glysosylases, family of base excision repair enzymes that show preference for both oxidative purines and pyrimidine lesions. For a description of all known DNA glycosylases and their families see the wikepedia page on DNA glycosylases [[1]]. Originally identified through in-silico studies from Wallace, Mitra and Seeger group back in 2002[1],[2],[3] the crystallisation of NEIL3 remained challenge for almost a decade due to the difficulty in getting a stabilized form of the protein for crystallization until a truncated version was crystallized in 2013 [4] and the crystal structure publlished as .
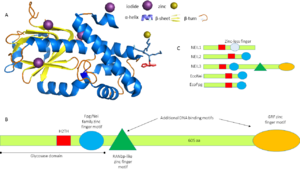
NEIL3 proteins are almost twice the size of other Fpg/Nei family members (Figure 1C). This protein belongs to alpha/beta[[2]] category of proteins with the following secondary structure elements: , , , , , ,,,,, , ,,,, . The beta strands form a two-layered at the N-terminus in which each layer is composed of four anti-parallel beta strands. The C terminus has a Helix 2 turn Helix (H2TH) motif formed by and a canonical formed by the interconnecting loop between beta strands-9 and 10. In contrast to other Fpg/nei members, NEIL3 has two additional DNA binding domains at their C-terminus: one Ran binding protein (RanBP2)-type zinc finger motif and two identical GRF-zinc finger motif. The catalytic residue of MmuNEILL3 is a rather than a proline as for other Fpg/nei members but serves the same purpose in catalysis and is surrounded by that forms the DNA binding cleft and lesion binding pocket of NEIL3.
Function
MmuNeil3 is a bifunctional DNA glycosylase that exhibits a broad substrate recognition spectrum (Sp, Gh, FapyG, FapyA, MeFapyG, DHU, DHT, 5-OHU, 5-OHC,5-OHMH, Tg, 8-oxoA, AP) but not 8-oxo-G. Image:Dna lesions.jpg
You may include any references to papers as in: the use of JSmol in Proteopedia [5] or to the article describing Jmol [6] to the rescue.
Function
== Disease
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst). 2002 Jul 17;1(7):517-29. PMID:12509226
- ↑ Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3523-8. PMID:11904416 doi:http://dx.doi.org/10.1073/pnas.062053799
- ↑ Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002 Nov 15;30(22):4926-36. PMID:12433996
- ↑ Liu M, Imamura K, Averill AM, Wallace SS, Doublie S. Structural Characterization of a Mouse Ortholog of Human NEIL3 with a Marked Preference for Single-Stranded DNA. Structure. 2013 Feb 5;21(2):247-56. doi: 10.1016/j.str.2012.12.008. Epub 2013 Jan, 9. PMID:23313161 doi:http://dx.doi.org/10.1016/j.str.2012.12.008
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
