We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Brittany Carroll/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 37: | Line 37: | ||
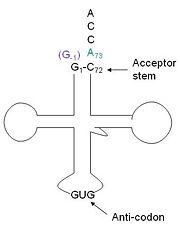
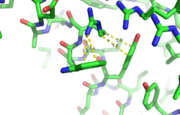
Hydrogen bonds between N’ and the backbone of N3 and N3 with N’ backbone are shown in the figure. The figure is difficult to see the T with P bb but it is not linear, this may just be due to modeling as it is close enough to form a h-bond. There is also a hydrophobic interaction between P and L. | Hydrogen bonds between N’ and the backbone of N3 and N3 with N’ backbone are shown in the figure. The figure is difficult to see the T with P bb but it is not linear, this may just be due to modeling as it is close enough to form a h-bond. There is also a hydrophobic interaction between P and L. | ||
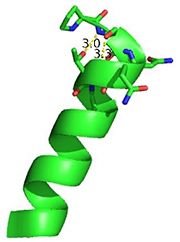
| - | [[Image:Cationpi.png|300|Right|thumb|This image depicts two cation-π interactions between Arg and Tyr or Trp. The energetic significances are -1.22 and -6.55 kj/mol respectively. | + | [[Image:Cationpi.png|300|Right|thumb|This image depicts two cation-π interactions between Arg and Tyr or Trp. The energetic significances are -1.22 and -6.55 kj/mol respectively. <ref>http://www.capture.caltech.edu/</ref> [[3otb]]]] |
A [http://www.en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interaction] occurs between a cation and the face of a simple aromatic, there is partial negative charge in the center of the ring. The cation-π interaction is actually stronger than a salt bridge because of the desolvation penalty. With the cation-π interaction the cation has a similar dosolvation penalty to pay as the salt bridge ions but the π system is already poorly solvated. Also there is not neutralization of charge that occurs between the two groups. These properties of the cation-π interaction imply that thecation-π interactions on protein surfaces (mainly where they are seen) could contribute to protein structure and stability. | A [http://www.en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interaction] occurs between a cation and the face of a simple aromatic, there is partial negative charge in the center of the ring. The cation-π interaction is actually stronger than a salt bridge because of the desolvation penalty. With the cation-π interaction the cation has a similar dosolvation penalty to pay as the salt bridge ions but the π system is already poorly solvated. Also there is not neutralization of charge that occurs between the two groups. These properties of the cation-π interaction imply that thecation-π interactions on protein surfaces (mainly where they are seen) could contribute to protein structure and stability. | ||
Revision as of 00:52, 28 April 2014
tRNA(His) guanylyltransferase
| |||||||||||
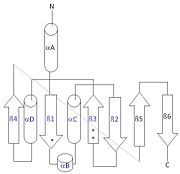
Addition SD Structures of Thg1
3otc, 3otd, 3ote - Thg1 - Homo sapiens
4kgk, 4kgm - Thg1-like - Bacillus thuringiensis
3wbz, 3wc0, 3wc1, 3wc2 - Thg1 - Candida albicans
References
- ↑ Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. PMID:22456265 doi:http://dx.doi.org/10.1261/rna.032300.112
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Rao BS, Eckenroth BE, Jackman JE, Doublie S. Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites. PLoS One. 2013 Jul 3;8(7):e67465. doi: 10.1371/journal.pone.0067465. Print 2013. PMID:23844012 doi:10.1371/journal.pone.0067465
- ↑ Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. PMID:22456265 doi:http://dx.doi.org/10.1261/rna.032300.112
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci U S A. 2010 Nov 8. PMID:21059936 doi:10.1073/pnas.1010436107
- ↑ Nakamura A, Nemoto T, Heinemann IU, Yamashita K, Sonoda T, Komoda K, Tanaka I, Soll D, Yao M. Structural basis of reverse nucleotide polymerization. Proc Natl Acad Sci U S A. 2013 Dec 24;110(52):20970-5. doi:, 10.1073/pnas.1321312111. Epub 2013 Dec 9. PMID:24324136 doi:http://dx.doi.org/10.1073/pnas.1321312111
- ↑ http://www.capture.caltech.edu/