1bh0
From Proteopedia
| Line 7: | Line 7: | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1bh0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1bh0 OCA], [http://www.ebi.ac.uk/pdbsum/1bh0 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1bh0 RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
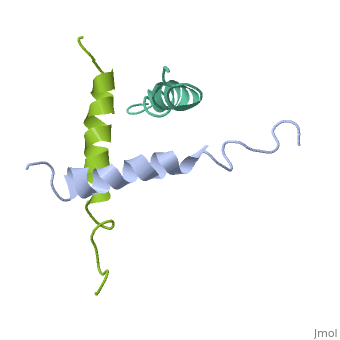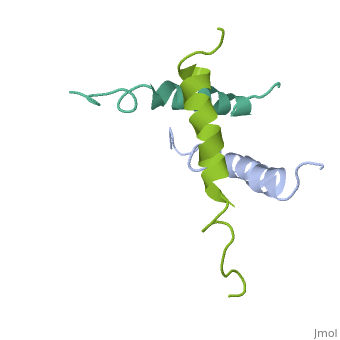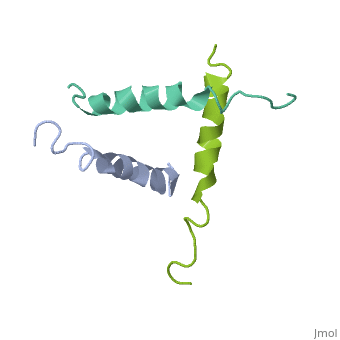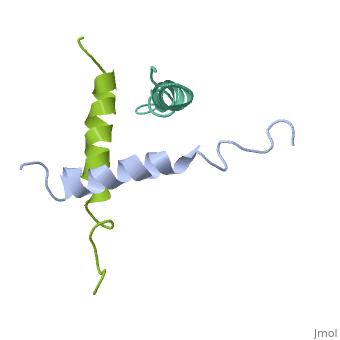
We have designed and synthesized eight compounds 2-9 which incorporate various amino acid residues in positions 17, 18, and 21 of the glucagon molecule: 2, [Lys17]glucagon amide; 3, [Lys18]glucagon amide; 4, [Nle17,Lys18,Glu21]glucagon amide; 5, [Orn17,18, Glu21]glucagon amide; 6, [d-Arg17]glucagon; 7, [d-Arg18]glucagon; 8, [d-Phe17]glucagon; and 9, [d-Phe18]glucagon. Compared to glucagon (IC50 = 1.5 nM), analogues 2-9 were found to have binding affinity IC50 values (in nM) of 0.7, 4.1, 1.0, 2.0, 5.0, 25.0, 43.0, and 32.0, respectively. When these compounds were tested for their ability to stimulate adenylate cyclase (AC) activity, they were found to be full or partial agonists having maximum stimulation values of 100, 100, 100, 100, 87, 78, 94, and 100%, respectively. On the basis of the X-ray crystal structure of [Lys17,18,Glu21]glucagon amide reported here, the ability to form a salt bridge between Lys18 and Glu21 is probably key to their increased binding and second messenger activities. Among the eight analogues synthesized here, only analogue 4 preserves the ability to form a salt bridge between Lys18 and Glu21. However, since these modifications are minor they do not seem to change the amphiphilic character of the C-terminus, allowing these analogues to reach 78-100% stimulation in the adenylate cyclase assay. Biological data from analogues 6-9 supports the idea that position 18 of glucagon may influence binding only, while position 17 may influence both receptor recognition and transduction. | We have designed and synthesized eight compounds 2-9 which incorporate various amino acid residues in positions 17, 18, and 21 of the glucagon molecule: 2, [Lys17]glucagon amide; 3, [Lys18]glucagon amide; 4, [Nle17,Lys18,Glu21]glucagon amide; 5, [Orn17,18, Glu21]glucagon amide; 6, [d-Arg17]glucagon; 7, [d-Arg18]glucagon; 8, [d-Phe17]glucagon; and 9, [d-Phe18]glucagon. Compared to glucagon (IC50 = 1.5 nM), analogues 2-9 were found to have binding affinity IC50 values (in nM) of 0.7, 4.1, 1.0, 2.0, 5.0, 25.0, 43.0, and 32.0, respectively. When these compounds were tested for their ability to stimulate adenylate cyclase (AC) activity, they were found to be full or partial agonists having maximum stimulation values of 100, 100, 100, 100, 87, 78, 94, and 100%, respectively. On the basis of the X-ray crystal structure of [Lys17,18,Glu21]glucagon amide reported here, the ability to form a salt bridge between Lys18 and Glu21 is probably key to their increased binding and second messenger activities. Among the eight analogues synthesized here, only analogue 4 preserves the ability to form a salt bridge between Lys18 and Glu21. However, since these modifications are minor they do not seem to change the amphiphilic character of the C-terminus, allowing these analogues to reach 78-100% stimulation in the adenylate cyclase assay. Biological data from analogues 6-9 supports the idea that position 18 of glucagon may influence binding only, while position 17 may influence both receptor recognition and transduction. | ||
| - | |||
| - | ==Disease== | ||
| - | Known disease associated with this structure: Hyperproglucagonemia OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=138030 138030]] | ||
==About this Structure== | ==About this Structure== | ||
| Line 34: | Line 34: | ||
[[Category: synthetic hormone]] | [[Category: synthetic hormone]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 19:00:46 2008'' |
Revision as of 16:00, 30 March 2008
| |||||||
| , resolution 3.0Å | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
STRUCTURE OF A GLUCAGON ANALOG
Overview
We have designed and synthesized eight compounds 2-9 which incorporate various amino acid residues in positions 17, 18, and 21 of the glucagon molecule: 2, [Lys17]glucagon amide; 3, [Lys18]glucagon amide; 4, [Nle17,Lys18,Glu21]glucagon amide; 5, [Orn17,18, Glu21]glucagon amide; 6, [d-Arg17]glucagon; 7, [d-Arg18]glucagon; 8, [d-Phe17]glucagon; and 9, [d-Phe18]glucagon. Compared to glucagon (IC50 = 1.5 nM), analogues 2-9 were found to have binding affinity IC50 values (in nM) of 0.7, 4.1, 1.0, 2.0, 5.0, 25.0, 43.0, and 32.0, respectively. When these compounds were tested for their ability to stimulate adenylate cyclase (AC) activity, they were found to be full or partial agonists having maximum stimulation values of 100, 100, 100, 100, 87, 78, 94, and 100%, respectively. On the basis of the X-ray crystal structure of [Lys17,18,Glu21]glucagon amide reported here, the ability to form a salt bridge between Lys18 and Glu21 is probably key to their increased binding and second messenger activities. Among the eight analogues synthesized here, only analogue 4 preserves the ability to form a salt bridge between Lys18 and Glu21. However, since these modifications are minor they do not seem to change the amphiphilic character of the C-terminus, allowing these analogues to reach 78-100% stimulation in the adenylate cyclase assay. Biological data from analogues 6-9 supports the idea that position 18 of glucagon may influence binding only, while position 17 may influence both receptor recognition and transduction.
About this Structure
1BH0 is a Single protein structure of sequence from [1]. Full crystallographic information is available from OCA.
Reference
Structure-function studies on positions 17, 18, and 21 replacement analogues of glucagon: the importance of charged residues and salt bridges in glucagon biological activity., Sturm NS, Lin Y, Burley SK, Krstenansky JL, Ahn JM, Azizeh BY, Trivedi D, Hruby VJ, J Med Chem. 1998 Jul 16;41(15):2693-700. PMID:9667960
Page seeded by OCA on Sun Mar 30 19:00:46 2008