1buy
From Proteopedia
| Line 7: | Line 7: | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1buy FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1buy OCA], [http://www.ebi.ac.uk/pdbsum/1buy PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1buy RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
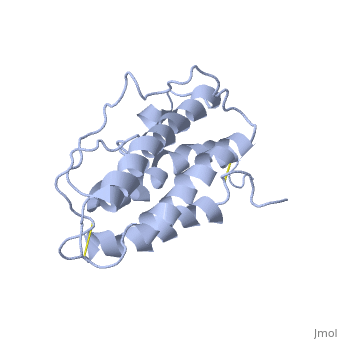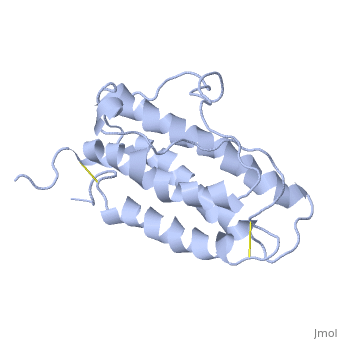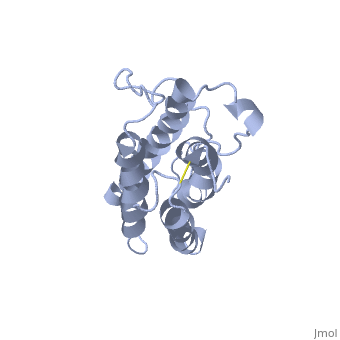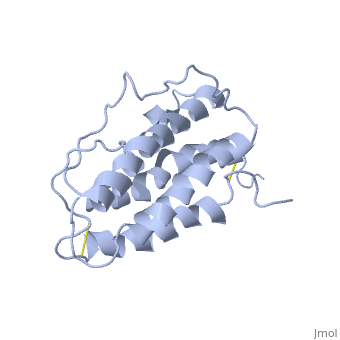
The solution structure of human erythropoietin (EPO) has been determined by nuclear magnetic resonance spectroscopy and the overall topology of the protein is revealed as a novel combination of features taken from both the long-chain and short-chain families of hematopoietic growth factors. Using the structure and data from mutagenesis studies we have elucidated the key physiochemical properties defining each of the two receptor binding sites on the EPO protein. A comparison of the NMR structure of the free EPO ligand to the receptor bound form, determined by X-ray crystallography, reveals conformational changes that may accompany receptor binding. | The solution structure of human erythropoietin (EPO) has been determined by nuclear magnetic resonance spectroscopy and the overall topology of the protein is revealed as a novel combination of features taken from both the long-chain and short-chain families of hematopoietic growth factors. Using the structure and data from mutagenesis studies we have elucidated the key physiochemical properties defining each of the two receptor binding sites on the EPO protein. A comparison of the NMR structure of the free EPO ligand to the receptor bound form, determined by X-ray crystallography, reveals conformational changes that may accompany receptor binding. | ||
| - | |||
| - | ==Disease== | ||
| - | Known disease associated with this structure: Erythremia (1) OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=133170 133170]] | ||
==About this Structure== | ==About this Structure== | ||
| Line 37: | Line 37: | ||
[[Category: growth factor]] | [[Category: growth factor]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 19:08:48 2008'' |
Revision as of 16:08, 30 March 2008
| |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
HUMAN ERYTHROPOIETIN, NMR MINIMIZED AVERAGE STRUCTURE
Overview
The solution structure of human erythropoietin (EPO) has been determined by nuclear magnetic resonance spectroscopy and the overall topology of the protein is revealed as a novel combination of features taken from both the long-chain and short-chain families of hematopoietic growth factors. Using the structure and data from mutagenesis studies we have elucidated the key physiochemical properties defining each of the two receptor binding sites on the EPO protein. A comparison of the NMR structure of the free EPO ligand to the receptor bound form, determined by X-ray crystallography, reveals conformational changes that may accompany receptor binding.
About this Structure
1BUY is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
NMR structure of human erythropoietin and a comparison with its receptor bound conformation., Cheetham JC, Smith DM, Aoki KH, Stevenson JL, Hoeffel TJ, Syed RS, Egrie J, Harvey TS, Nat Struct Biol. 1998 Oct;5(10):861-6. PMID:9783743
Page seeded by OCA on Sun Mar 30 19:08:48 2008