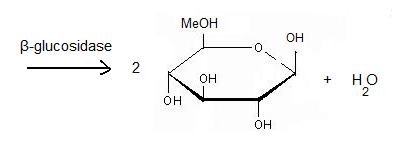
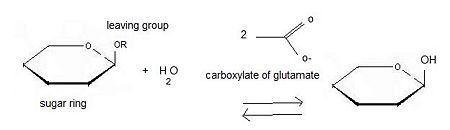
Beta-glucosidase
From Proteopedia
| Line 1: | Line 1: | ||
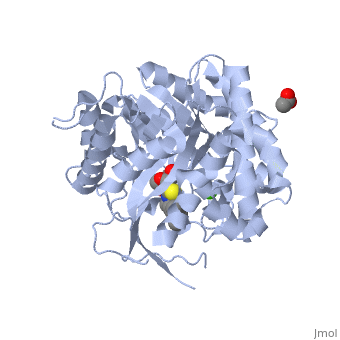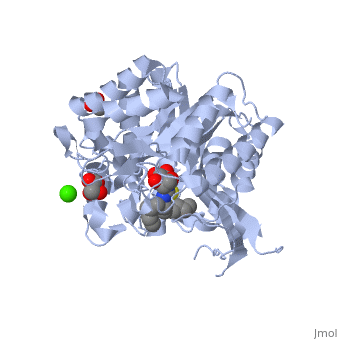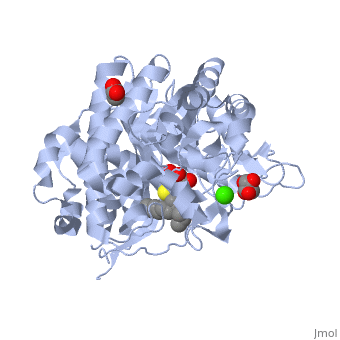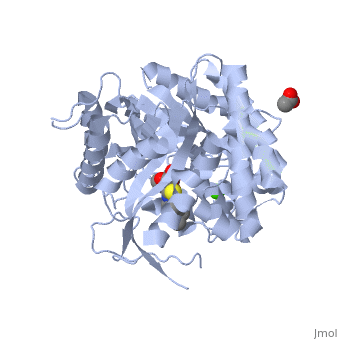
<StructureSection load='2vrj' size='450' side='right' scene='' caption='β-glucosidase complex with calystegine analog, acetate and Ca+2 ion (green) (PDB code [[2vrj]])'> | <StructureSection load='2vrj' size='450' side='right' scene='' caption='β-glucosidase complex with calystegine analog, acetate and Ca+2 ion (green) (PDB code [[2vrj]])'> | ||
| - | + | ||
{{Clear}} | {{Clear}} | ||
'''β-glucosidase''' is an enzyme which catalyses the hydrolysis of terminal non-reducing residues in β-glucosides (EC number : 3.2.1.21). In the case of 2VRJ, it comes from ''Thermotoga maritima'' which is a rod-shaped bacterium belonging to the order of Thermotogates. This bacterium was originally isolated from geothermal heated marine sediments. | '''β-glucosidase''' is an enzyme which catalyses the hydrolysis of terminal non-reducing residues in β-glucosides (EC number : 3.2.1.21). In the case of 2VRJ, it comes from ''Thermotoga maritima'' which is a rod-shaped bacterium belonging to the order of Thermotogates. This bacterium was originally isolated from geothermal heated marine sediments. | ||
Revision as of 10:10, 25 August 2014
| |||||||||||
Contents |
3D structures of Beta-glucosidase
Updated on 25-August-2014
3ahx – GBA – Clostridium cellulovorans
3ahy – TrGB – Trichoderma reesei
3abz – KmGB – Kluyveromyces marxianus
2x40 – TnGB3B – Thermotoga neapolitana
3gno - JrGB residues 38-521 – Japanese rice
2rgl, 2rgm - JrGB residues 29-504
3f4v, 3f5j, 3f5k, 3f5l,3scn, 3sco, 3sct, 3scu, 3scv, 3scw, 3scp, 3scr - JrGB7 (mutant)
3ahz – tGB – termite
3fiy, 3cmj - UbGB catalytic domain (mutant) – Uncultured bacteria
2o9p, 1bga – PpGBB – Paenibacillus polymyxa
2jfe, 3gxd, 3gxi, 3gxm - hGB cystolic – human
3ke0, 3keh - hGB (mutant)
2e3z – PcGB – Phanerochaete crysosporium
2dga - wGB residues 1-520 – wheat
2vff, 1vff – GB – Pyrococcus horikoshii
1oif, 1od0 – TmGB catalytic domain - Thermotoga maritima
4gxp – TmGB/TrGB
1ug6 – GB - Thermus thermophilus
1hxj, 1e1e – ZmGB – Zea mays
1e4l - ZmGB (mutant)
1gon – SsGB – Streptomyces sp.
1qox – GB - Bacillus circulans
1tr1 - BpGBB (mutant) - Bacillus polymyxa
1cbg – GB cyanogenic – White clover
3aiu - rGB residues 50-568 – rye
3f93, 3f94 – PsGB residues 28-840 – Pseudoalteromonas
3f95 – PsGB residues 657-840
3ptk – rGB - rice
3apg – GB – Pyrococcus furiosus
3ta9 – GB – Halothermothrix orenii
3zyz, 3zz1 – GB – Hypocrea jecorina
4hz6 – ubGB – uncultured bacterium
4bce – GB (mutant) – Thermus thermophilus
4iib – AaGB – Aspergillus aculeatus
3w53 – GB – Micrococcus antarcticus
Beta-glucosidase complex with sugar
3ptm, 3ptq – rGB + glucoside
3wba, 3wbe – rGB (mutant) + glucose
3ai0 – tGB + glucoside
3ac0 - KmGB + glucoside
2x41, 2x42 - TnGB + glucoside
3air – wGB residues 50-569 + glucoside + dinitrophenol
3ais - wGB residues 50-569 (mutant) + glucoside + aglycone
3aiq - wGB residues 50-569 + aglycone
3aiv - rGB residues 50-568 + aglycone
3aiw - rGB residues 50-568 + glucoside + dinitrophenol
3gnp, 3gnr - JrGB residues 38-521 + glucoside
3aht, 3ahv, 3scq, 3scs – JrGB7 (mutant) + saccharide
1oin - TmGBA + glucoside
3fiz, 3fj0 - UbGB residues 18-482 (mutant) + glucoside
2zox, 2e9l, 2e9m - hGB cystolic + glucoside
2o9s, 2o9r, 2z1s – PpGB + saccharide
2o9t - PpGB + glucoside
1uyq - PpGB (mutant) + glucoside
1bgg - PpGB + gluconate
2jie - BpGB + glucoside
1e4i - BpGB (mutant) + glucoside
2e40 – PcGB + gluconolactone
3vif - NkGB + gluconolactone – Neotermes koshunensis
3vig, 3vii - NkGB + saccharide
3vih – NkGB + glycerol
3vij – NkGB (mutant) + glucose
3vik, 3vil, 3vim, 3vin, 3vio, 3vip – NkGB (mutant) + saccharide
4hz7, 4hz8 - ubGB (mutant) + glucose
1v08 – ZmGB + gluco-tetrazole
1e1f - ZmGB + glucoside
1h49, 1e4n, 1e56 - ZmGB (mutant) + aglycone
1gnx - SsGB + saccharide - Sulfolobus solfataricus
3gfx – hGB + drug
4iic, 4iid, 4iie, 4iif – AaGB + drug
4iig, 4iih – AaGB + saccharide
3vkk – hGB + mannose
4i3g – GB + glucose – Streptomyces venezuelae
Beta-glucosidase complex with inhibitor
2wbg, 2wc3, 2wc4, 2vrj, 2jal, 2j75, 2j77, 2j78, 2j79, 2j7b, 2j7d, 2j7e, 2j7f, 2j7g, 2j7h, 2j7c, 2ces, 2cet, 2cbu, 2cbv, 1uz1, 1w3j, 1oim – TmGBA + inhibitor
2cer – SsGB + inhibitor
1e55 - ZmGB (mutant) + inhibitor
3rik, 3ril - hGB + inhibitor
6-phospho-β-glucosidase
1s6y – PGB – Geobacillus stearothermophilus
1up4 – TmPGB
1up6, 1up7 – TmPGB + NAD + G6P
3qom, 4gze – PGB – Lactobacillus plantarum
2xhy – PGB – Escherichia coli
1h4p – PGB I/II – yeast
3eqn – WfPGB – White-rot fungus
3eqo – WfPGB + glucolactone
4b3k, 4b3l – GB – Streptococcus pyogenes
4gpn – GB + gentiobiose 6-phosphate – Streptococcus mutans
4ipl – SpGB – Streptococcus pneumoniae
4ipn – SpGB + thiocellobiose
Glucan 1,3-β-glucosidase
3n9k – CaGGB + glucoside – Candida albicans
3o6a – CaGGB (mutant)
3ur7, 3ur8 – poGGB – potato
4gzi – poGGB (mutant)
4gzj - poGGB (mutant) + saccharide
Raucaffricine-β-glucosidase
3u57, 3u5u – dpRGB (mutant) – devilpepper
3u5y – dpRGB (mutant)m + secologanin
4a3y, 4atd – seRGB - serpentwood
4atl – seRGB + glucose
4ek7 – seRGB (mutant)
3zj6 – seRGB + inhibitor
Strictosidine-β-glucosidase
3zj7, 3zj8 – seSGB + inhibitor
References
- ↑ Aguilar M, Gloster TM, Garcia-Moreno MI, Ortiz Mellet C, Davies GJ, Llebaria A, Casas J, Egido-Gabas M, Garcia Fernandez JM. Molecular basis for beta-glucosidase inhibition by ring-modified calystegine analogues. Chembiochem. 2008 Nov 3;9(16):2612-8. PMID:18833549 doi:10.1002/cbic.200800451
- ↑ http://en.wikipedia.org/wiki/B-glucosidase
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.ebi.ac.uk/interpro/IEntry?ac=IPR018120#PUB00002205
- ↑ http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/CSA/CSA_Site_Wrapper.pl?pdb=2vrj
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.cazy.org/fam/ghf_INV_RET.html#3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Muriel Breteau, Alexander Berchansky, Joel L. Sussman, David Canner