1f8a
From Proteopedia
| Line 4: | Line 4: | ||
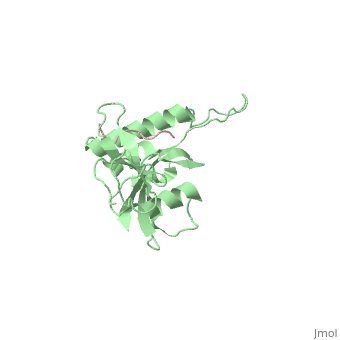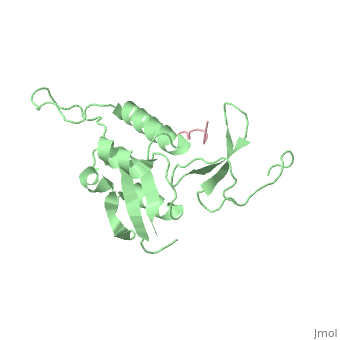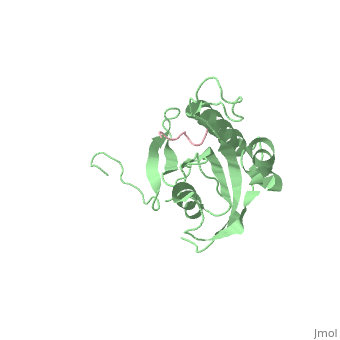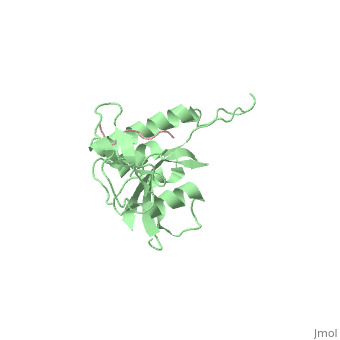
|PDB= 1f8a |SIZE=350|CAPTION= <scene name='initialview01'>1f8a</scene>, resolution 1.84Å | |PDB= 1f8a |SIZE=350|CAPTION= <scene name='initialview01'>1f8a</scene>, resolution 1.84Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= | + | |LIGAND= <scene name='pdbligand=SEP:PHOSPHOSERINE'>SEP</scene> |
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Peptidylprolyl_isomerase Peptidylprolyl isomerase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=5.2.1.8 5.2.1.8] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Peptidylprolyl_isomerase Peptidylprolyl isomerase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=5.2.1.8 5.2.1.8] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[1pin|1PIN]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1f8a FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1f8a OCA], [http://www.ebi.ac.uk/pdbsum/1f8a PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1f8a RCSB]</span> | ||
}} | }} | ||
| Line 32: | Line 35: | ||
[[Category: ww domain]] | [[Category: ww domain]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 20:17:51 2008'' |
Revision as of 17:17, 30 March 2008
| |||||||
| , resolution 1.84Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Activity: | Peptidylprolyl isomerase, with EC number 5.2.1.8 | ||||||
| Related: | 1PIN
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS
Overview
Pin1 contains an N-terminal WW domain and a C-terminal peptidyl-prolyl cis-trans isomerase (PPIase) domain connected by a flexible linker. To address the energetic and structural basis for WW domain recognition of phosphoserine (P.Ser)/phosphothreonine (P. Thr)- proline containing proteins, we report the energetic and structural analysis of a Pin1-phosphopeptide complex. The X-ray crystal structure of Pin1 bound to a doubly phosphorylated peptide (Tyr-P.Ser-Pro-Thr-P.Ser-Pro-Ser) representing a heptad repeat of the RNA polymerase II large subunit's C-terminal domain (CTD), reveals the residues involved in the recognition of a single P.Ser side chain, the rings of two prolines, and the backbone of the CTD peptide. The side chains of neighboring Arg and Ser residues along with a backbone amide contribute to recognition of P.Ser. The lack of widespread conservation of the Arg and Ser residues responsible for P.Ser recognition in the WW domain family suggests that only a subset of WW domains can bind P.Ser-Pro in a similar fashion to that of Pin1.
About this Structure
1F8A is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Structural basis for phosphoserine-proline recognition by group IV WW domains., Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP, Nat Struct Biol. 2000 Aug;7(8):639-43. PMID:10932246
Page seeded by OCA on Sun Mar 30 20:17:51 2008