We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1z7n
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Structural highlights == | == Structural highlights == | ||
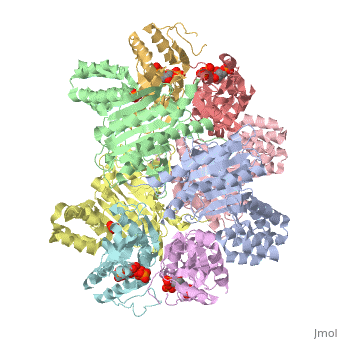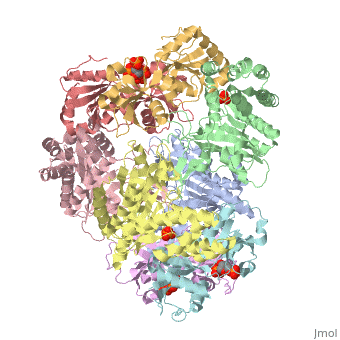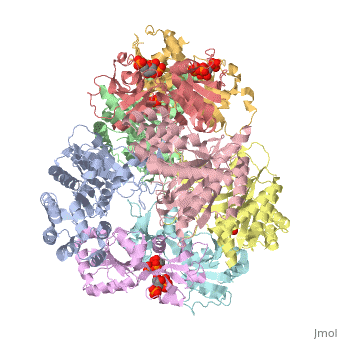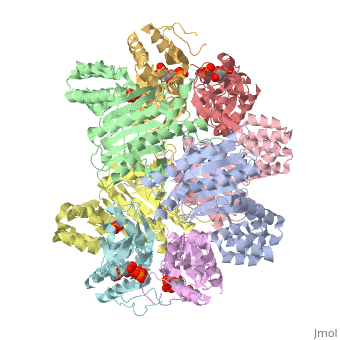
<table><tr><td colspan='2'>[[1z7n]] is a 8 chain structure with sequence from [http://en.wikipedia.org/wiki/Lactococcus_lactis Lactococcus lactis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Z7N OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1Z7N FirstGlance]. <br> | <table><tr><td colspan='2'>[[1z7n]] is a 8 chain structure with sequence from [http://en.wikipedia.org/wiki/Lactococcus_lactis Lactococcus lactis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Z7N OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1Z7N FirstGlance]. <br> | ||
| - | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=PO4:PHOSPHATE+ION'>PO4</scene>, <scene name='pdbligand=PRP:ALPHA-PHOSPHORIBOSYLPYROPHOSPHORIC+ACID'>PRP</scene>< | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=PO4:PHOSPHATE+ION'>PO4</scene>, <scene name='pdbligand=PRP:ALPHA-PHOSPHORIBOSYLPYROPHOSPHORIC+ACID'>PRP</scene></td></tr> |
| - | <tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1z7m|1z7m]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1z7m|1z7m]]</td></tr> |
| - | <tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">hisZ ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=1358 Lactococcus lactis]), hisG ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=1358 Lactococcus lactis])</td></tr> | + | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">hisZ ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=1358 Lactococcus lactis]), hisG ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=1358 Lactococcus lactis])</td></tr> |
| - | <tr><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/ATP_phosphoribosyltransferase ATP phosphoribosyltransferase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.4.2.17 2.4.2.17] </span></td></tr> | + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/ATP_phosphoribosyltransferase ATP phosphoribosyltransferase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.4.2.17 2.4.2.17] </span></td></tr> |
| - | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1z7n FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1z7n OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1z7n RCSB], [http://www.ebi.ac.uk/pdbsum/1z7n PDBsum]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1z7n FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1z7n OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1z7n RCSB], [http://www.ebi.ac.uk/pdbsum/1z7n PDBsum]</span></td></tr> |
| - | <table> | + | </table> |
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/HISZ_LACLA HISZ_LACLA]] Required for the first step of histidine biosynthesis. May allow the feedback regulation of ATP phosphoribosyltransferase activity by histidine.[HAMAP-Rule:MF_00125] [[http://www.uniprot.org/uniprot/HIS1_LACLA HIS1_LACLA]] Catalyzes the condensation of ATP and 5-phosphoribose 1-diphosphate to form N'-(5'-phosphoribosyl)-ATP (PR-ATP). Has a crucial role in the pathway because the rate of histidine biosynthesis seems to be controlled primarily by regulation of HisG enzymatic activity.[HAMAP-Rule:MF_01018] | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 36: | Line 38: | ||
[[Category: ATP phosphoribosyltransferase]] | [[Category: ATP phosphoribosyltransferase]] | ||
[[Category: Lactococcus lactis]] | [[Category: Lactococcus lactis]] | ||
| - | [[Category: Champagne, K S | + | [[Category: Champagne, K S]] |
| - | [[Category: Doublie, S | + | [[Category: Doublie, S]] |
| - | [[Category: Francklyn, C S | + | [[Category: Francklyn, C S]] |
| - | [[Category: Larrabee, Y | + | [[Category: Larrabee, Y]] |
| - | [[Category: Sissler, M | + | [[Category: Sissler, M]] |
[[Category: Allosteric]] | [[Category: Allosteric]] | ||
[[Category: Atp-prt]] | [[Category: Atp-prt]] | ||
Revision as of 23:15, 24 December 2014
ATP Phosphoribosyl transferase (HisZG ATP-PRTase) from Lactococcus lactis with bound PRPP substrate
| |||||||||||