We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1sqe
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Structural highlights == | == Structural highlights == | ||
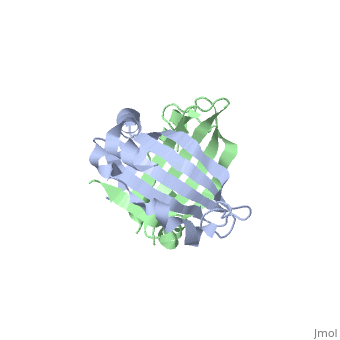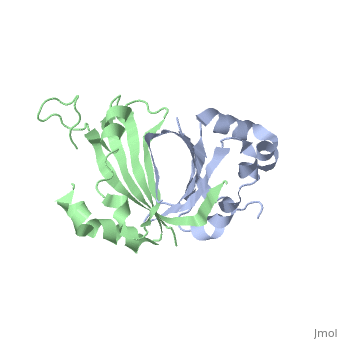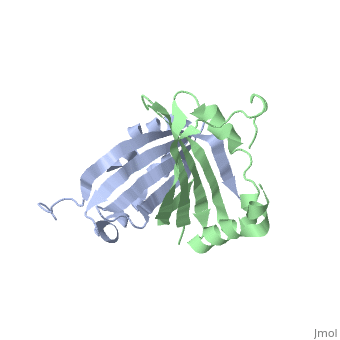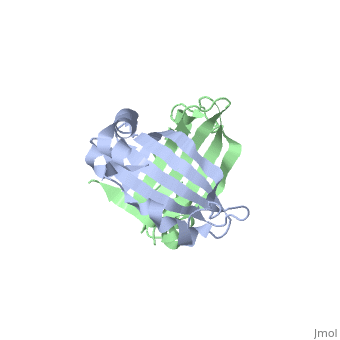
<table><tr><td colspan='2'>[[1sqe]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Staphylococcus_aureus Staphylococcus aureus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1SQE OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1SQE FirstGlance]. <br> | <table><tr><td colspan='2'>[[1sqe]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Staphylococcus_aureus Staphylococcus aureus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1SQE OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1SQE FirstGlance]. <br> | ||
| - | </td></tr><tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1sqe FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1sqe OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1sqe RCSB], [http://www.ebi.ac.uk/pdbsum/1sqe PDBsum], [http://www.topsan.org/Proteins/MCSG/1sqe TOPSAN]</span></td></tr> | + | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1sqe FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1sqe OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1sqe RCSB], [http://www.ebi.ac.uk/pdbsum/1sqe PDBsum], [http://www.topsan.org/Proteins/MCSG/1sqe TOPSAN]</span></td></tr> |
| - | <table> | + | </table> |
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/ISDI_STAAM ISDI_STAAM]] Allows bacterial pathogens to use the host heme as an iron source. Catalyzes the oxidative degradation of the heme macrocyclic porphyrin ring to the oxo-bilirubin chromophore staphylobilin (a mixture of the linear tetrapyrroles 5-oxo-delta-bilirubin and 15-oxo-beta-bilirubin) in the presence of a suitable electron donor such as ascorbate or NADPH--cytochrome P450 reductase, with subsequent release of free iron (By similarity).<ref>PMID:15520015</ref> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 31: | Line 33: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Staphylococcus aureus]] | [[Category: Staphylococcus aureus]] | ||
| - | [[Category: Joachimiak, A | + | [[Category: Joachimiak, A]] |
| - | [[Category: Joachimiak, G | + | [[Category: Joachimiak, G]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Category: Structural genomic]] | [[Category: Structural genomic]] | ||
| + | [[Category: Schneewind, O]] | ||
| + | [[Category: Wu, R]] | ||
| + | [[Category: Zhang, R]] | ||
| + | [[Category: Mcsg]] | ||
| + | [[Category: PSI, Protein structure initiative]] | ||
[[Category: Unknown function]] | [[Category: Unknown function]] | ||
Revision as of 09:03, 25 December 2014
1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics
| |||||||||||