1ls8
From Proteopedia
| Line 7: | Line 7: | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ls8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ls8 OCA], [http://www.ebi.ac.uk/pdbsum/1ls8 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1ls8 RCSB]</span> | ||
}} | }} | ||
| Line 34: | Line 37: | ||
[[Category: solution structure]] | [[Category: solution structure]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 22:06:41 2008'' |
Revision as of 19:06, 30 March 2008
| |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
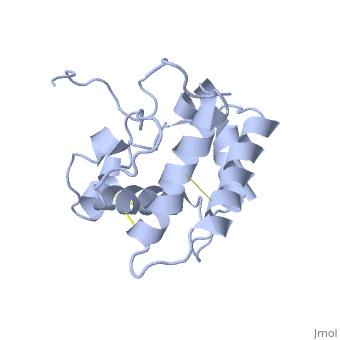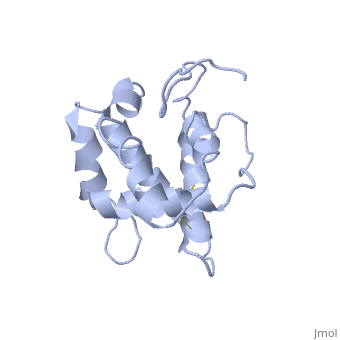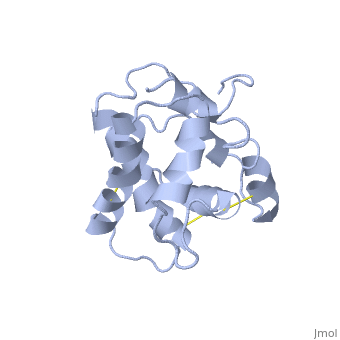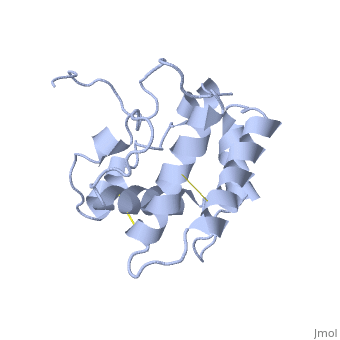
NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH
Overview
The nuclear magnetic resonance structure of the unliganded pheromone-binding protein (PBP) from Bombyx mori at pH above 6.5, BmPBP(B), consists of seven helices with residues 3-8, 16-22, 29-32, 46-59, 70-79, 84-100, and 107-124, and contains the three disulfide bridges 19-54, 50-108, and 97-117. This polypeptide fold encloses a large hydrophobic cavity, with a sufficient volume to accommodate the natural ligand bombykol. The polypeptide folds in free BmPBP(B) and in crystals of a BmPBP-bombykol complex are nearly identical, indicating that the B-form of BmPBP in solution represents the active conformation for ligand binding.
About this Structure
1LS8 is a Single protein structure of sequence from Bombyx mori. Full crystallographic information is available from OCA.
Reference
NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH., Lee D, Damberger FF, Peng G, Horst R, Guntert P, Nikonova L, Leal WS, Wuthrich K, FEBS Lett. 2002 Nov 6;531(2):314-8. PMID:12417333
Page seeded by OCA on Sun Mar 30 22:06:41 2008