Molecular Playground/E. coli ClpP
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== ClpP Introduction == | == ClpP Introduction == | ||
| - | ''E. coli'' Casein lytic proteinase P (ClpP) is a double ring [http://en.wiktionary.org/wiki/tetradecamer tetradecameric] homo oligomer compartmentalized peptidase [ | + | ''E. coli'' Casein lytic proteinase P (ClpP) is a double ring [http://en.wiktionary.org/wiki/tetradecamer tetradecameric] homo oligomer compartmentalized peptidase [http://www.ncbi.nlm.nih.gov/pubmed/?term=ClpP%3A+A+structurally+dynamic+protease+regulated+by+AAA%2B+proteins]. ClpP requires the use of ATP dependent regulatory elements that independently bind to ClpP in order for substrates to have access the active core [2-4]. Here, proteins that are translocated by regulatory elements into the peptidase core are cleaved into smaller amino acid chains approximately on average 6-8aa in length [5]. |
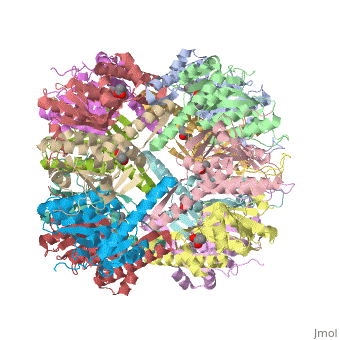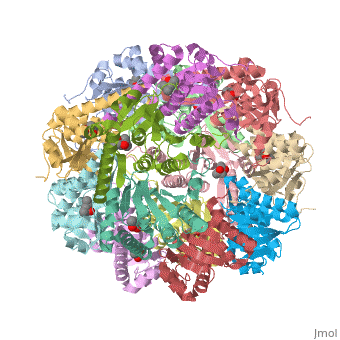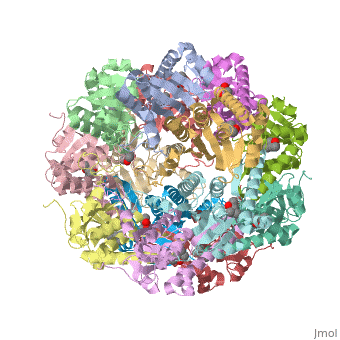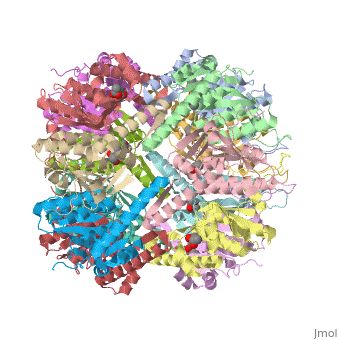
== Tetradecameric Structure == | == Tetradecameric Structure == | ||
ClpP is a serine protease which consists of fourteen monomers situated into two [http://en.wiktionary.org/wiki/heptamer heptameric] rings seated on top of each other. In the center of the barrel-shaped chamber lies a core of fourteen peptide-cleaving active sites, restricted by the narrow entrance called the ''axial pore''. As peptides are [http://en.wikipedia.org/wiki/Processivity processively] threaded in the pore by the regulatory elements, the peptides are then degraded into smaller fragments as a result from ClpP cleavage. Smaller peptides are then released through small openings found around the equatorial interface of the two stacked rings. | ClpP is a serine protease which consists of fourteen monomers situated into two [http://en.wiktionary.org/wiki/heptamer heptameric] rings seated on top of each other. In the center of the barrel-shaped chamber lies a core of fourteen peptide-cleaving active sites, restricted by the narrow entrance called the ''axial pore''. As peptides are [http://en.wikipedia.org/wiki/Processivity processively] threaded in the pore by the regulatory elements, the peptides are then degraded into smaller fragments as a result from ClpP cleavage. Smaller peptides are then released through small openings found around the equatorial interface of the two stacked rings. | ||
| Line 17: | Line 17: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | <ref>Alexopoulos JA et. al (2012 J Struct Bio)</ref> | + | <ref>1. Alexopoulos JA et. al (2012 J Struct Bio)</ref> |
== Acknowledgements== | == Acknowledgements== | ||
Kamal Joshi, Joanne Lau, Jing Liu, Rob Vass | Kamal Joshi, Joanne Lau, Jing Liu, Rob Vass | ||
Revision as of 20:07, 3 December 2014
Here in the Chien lab, we study how proteolysis plays a large part in protein quality control. The maintenance and timely destruction of protein levels plays an important role during cell homeostasis and cell transitions/differentiation, yet much of what governs these processes has yet to be fully understood.
| |||||||||||
References
Acknowledgements
Kamal Joshi, Joanne Lau, Jing Liu, Rob Vass