Apurinic-Apyrimidinic Endonuclease
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='2o3c' size='350' side='right' caption='Figure 2. Zebrafish APE1 trimer interacting with Pb+2 ions. [[2o3c]]' scene=''> | <StructureSection load='2o3c' size='350' side='right' caption='Figure 2. Zebrafish APE1 trimer interacting with Pb+2 ions. [[2o3c]]' scene=''> | ||
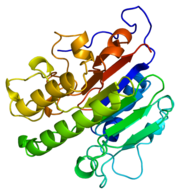
[[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | [[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | ||
| + | |||
| + | ==Function== | ||
| + | |||
DNA is damaged frequently in living cells via endogenous and exogenous factors that have the ability to cause abasic sites in the DNA strand. Abasic DNA must be repaired both accuratly and efficiently in order to prevent the death of the cell or the accumulation of mutations within the genome. Abasic DNA is repaired in the base excision repair pathway. '''Apurinic/apyrimidinic endonuclease-1''' (APE1) is the major repair enzyme in the base excision repair pathway in humans <ref name="Masood">Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879</ref>. APE1 has the ability to cleave the phophodiester bond at the 5' end of abasic sites in damaged DNA strands <ref name="Masood"/>. | DNA is damaged frequently in living cells via endogenous and exogenous factors that have the ability to cause abasic sites in the DNA strand. Abasic DNA must be repaired both accuratly and efficiently in order to prevent the death of the cell or the accumulation of mutations within the genome. Abasic DNA is repaired in the base excision repair pathway. '''Apurinic/apyrimidinic endonuclease-1''' (APE1) is the major repair enzyme in the base excision repair pathway in humans <ref name="Masood">Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879</ref>. APE1 has the ability to cleave the phophodiester bond at the 5' end of abasic sites in damaged DNA strands <ref name="Masood"/>. | ||
| Line 7: | Line 10: | ||
The oxidation states of molecules, such as many transcription factors, within the cell can activate or deactivate molecules and therefore regulate the expression of genes <ref name="Ando"> Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.</ref>The oxidation states of many amino acid residues within a protein can change the properties of the protein which can greatly affect the enzymatic activity of the protein. The oxidation states of cysteine residues, among other amino acid residues, within a polypeptide are regulated by APE1<ref name="Ando"/>.Proteins that APE1 has been established to regulate the oxidation states of the cysteine residues of include NF-kB, Ehr-1, HIF-1, HLF, Pax-5, Pax-8 and p53, the mechanism of how APE1 contributes to the oxidation states of the cysteine residues on these proteins, however, is poorly understood<ref name="Ando"/> It is due to these many functions of APE1 that APE1 has become known as the multifunctional workhorse protein of the cell. | The oxidation states of molecules, such as many transcription factors, within the cell can activate or deactivate molecules and therefore regulate the expression of genes <ref name="Ando"> Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.</ref>The oxidation states of many amino acid residues within a protein can change the properties of the protein which can greatly affect the enzymatic activity of the protein. The oxidation states of cysteine residues, among other amino acid residues, within a polypeptide are regulated by APE1<ref name="Ando"/>.Proteins that APE1 has been established to regulate the oxidation states of the cysteine residues of include NF-kB, Ehr-1, HIF-1, HLF, Pax-5, Pax-8 and p53, the mechanism of how APE1 contributes to the oxidation states of the cysteine residues on these proteins, however, is poorly understood<ref name="Ando"/> It is due to these many functions of APE1 that APE1 has become known as the multifunctional workhorse protein of the cell. | ||
| - | ==Structure | + | ==Structure== |
APE1's major biological role is its activity in the BER pathway. APE1 is classified as a DNA lyase that has a structure weight of 50000-75000Da, depending on the crystal structure in question <ref name="Mol">Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.</ref>. Many crystal structures of APE1 have been determined with amino acid numbers ranging from ~270 amino acid residues to ~320 amino acid residues<ref name="Mol"/><ref name="Masood"/>. The structure of APE1 has 15 anti-parallel beta sheets that line the interior of the protein, with 8 alpha helices lining the exterior of the protein. The beta sheets line the interior of the protein and form a convex binding surface for the DNA strand to fit in to. The binding surface of the protein is lined with positively charged amino acid residues that allow APE1 to bind to the negatively charged DNA strand. Metal ions assits the binding to the substrate by occupying the binding site. | APE1's major biological role is its activity in the BER pathway. APE1 is classified as a DNA lyase that has a structure weight of 50000-75000Da, depending on the crystal structure in question <ref name="Mol">Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.</ref>. Many crystal structures of APE1 have been determined with amino acid numbers ranging from ~270 amino acid residues to ~320 amino acid residues<ref name="Mol"/><ref name="Masood"/>. The structure of APE1 has 15 anti-parallel beta sheets that line the interior of the protein, with 8 alpha helices lining the exterior of the protein. The beta sheets line the interior of the protein and form a convex binding surface for the DNA strand to fit in to. The binding surface of the protein is lined with positively charged amino acid residues that allow APE1 to bind to the negatively charged DNA strand. Metal ions assits the binding to the substrate by occupying the binding site. | ||
Revision as of 10:53, 2 December 2015
| |||||||||||
3D structures of Apurinic/Apyrimidinic Endonuclease
Updated on 02-December-2015
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879
- ↑ 2.0 2.1 Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958
- ↑ 3.0 3.1 3.2 Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
Proteopedia Page Contributors and Editors (what is this?)
Mark Barnes, Michal Harel, Alexander Berchansky, David Canner, Jaime Prilusky, Andrea Gorrell, Eran Hodis, Joel L. Sussman