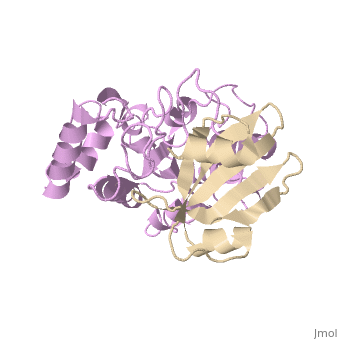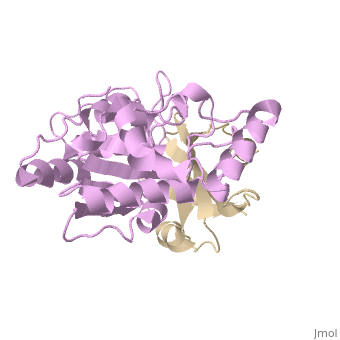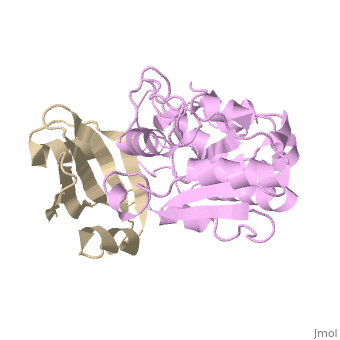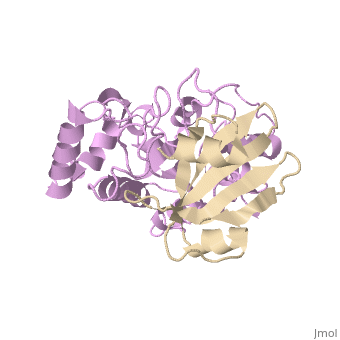1ugh
From Proteopedia
| Line 5: | Line 5: | ||
|SITE= | |SITE= | ||
|LIGAND= | |LIGAND= | ||
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Uridine_nucleosidase Uridine nucleosidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.2.2.3 3.2.2.3] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Uridine_nucleosidase Uridine nucleosidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.2.2.3 3.2.2.3] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ugh FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ugh OCA], [http://www.ebi.ac.uk/pdbsum/1ugh PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1ugh RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
Uracil-DNA glycosylase inhibitor (Ugi) is a B. subtilis bacteriophage protein that protects the uracil-containing phage DNA by irreversibly inhibiting the key DNA repair enzyme uracil-DNA glycosylase (UDG). The 1.9 A crystal structure of Ugi complexed to human UDG reveals that the Ugi structure, consisting of a twisted five-stranded antiparallel beta sheet and two alpha helices, binds by inserting a beta strand into the conserved DNA-binding groove of the enzyme without contacting the uracil specificity pocket. The resulting interface, which buries over 1200 A2 on Ugi and involves the entire beta sheet and an alpha helix, is polar and contains 22 water molecules. Ugi binds the sequence-conserved DNA-binding groove of UDG via shape and electrostatic complementarity, specific charged hydrogen bonds, and hydrophobic packing enveloping Leu-272 from a protruding UDG loop. The apparent mimicry by Ugi of DNA interactions with UDG provides both a structural mechanism for UDG binding to DNA, including the enzyme-assisted expulsion of uracil from the DNA helix, and a crystallographic basis for the design of inhibitors with scientific and therapeutic applications. | Uracil-DNA glycosylase inhibitor (Ugi) is a B. subtilis bacteriophage protein that protects the uracil-containing phage DNA by irreversibly inhibiting the key DNA repair enzyme uracil-DNA glycosylase (UDG). The 1.9 A crystal structure of Ugi complexed to human UDG reveals that the Ugi structure, consisting of a twisted five-stranded antiparallel beta sheet and two alpha helices, binds by inserting a beta strand into the conserved DNA-binding groove of the enzyme without contacting the uracil specificity pocket. The resulting interface, which buries over 1200 A2 on Ugi and involves the entire beta sheet and an alpha helix, is polar and contains 22 water molecules. Ugi binds the sequence-conserved DNA-binding groove of UDG via shape and electrostatic complementarity, specific charged hydrogen bonds, and hydrophobic packing enveloping Leu-272 from a protruding UDG loop. The apparent mimicry by Ugi of DNA interactions with UDG provides both a structural mechanism for UDG binding to DNA, including the enzyme-assisted expulsion of uracil from the DNA helix, and a crystallographic basis for the design of inhibitors with scientific and therapeutic applications. | ||
| - | |||
| - | ==Disease== | ||
| - | Known diseases associated with this structure: Immunodeficiency with hyper IgM, type 4 OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191525 191525]] | ||
==About this Structure== | ==About this Structure== | ||
| - | 1UGH is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/ | + | 1UGH is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Bacillus_phage_pbs2 Bacillus phage pbs2] and [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1UGH OCA]. |
==Reference== | ==Reference== | ||
Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA., Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA, Cell. 1995 Sep 8;82(5):701-8. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/7671300 7671300] | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA., Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA, Cell. 1995 Sep 8;82(5):701-8. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/7671300 7671300] | ||
| + | [[Category: Bacillus phage pbs2]] | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: Phage pbs1]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
[[Category: Uridine nucleosidase]] | [[Category: Uridine nucleosidase]] | ||
| Line 38: | Line 38: | ||
[[Category: glycosylase]] | [[Category: glycosylase]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 00:09:56 2008'' |
Revision as of 21:09, 30 March 2008
| |||||||
| , resolution 1.90Å | |||||||
|---|---|---|---|---|---|---|---|
| Activity: | Uridine nucleosidase, with EC number 3.2.2.3 | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA
Overview
Uracil-DNA glycosylase inhibitor (Ugi) is a B. subtilis bacteriophage protein that protects the uracil-containing phage DNA by irreversibly inhibiting the key DNA repair enzyme uracil-DNA glycosylase (UDG). The 1.9 A crystal structure of Ugi complexed to human UDG reveals that the Ugi structure, consisting of a twisted five-stranded antiparallel beta sheet and two alpha helices, binds by inserting a beta strand into the conserved DNA-binding groove of the enzyme without contacting the uracil specificity pocket. The resulting interface, which buries over 1200 A2 on Ugi and involves the entire beta sheet and an alpha helix, is polar and contains 22 water molecules. Ugi binds the sequence-conserved DNA-binding groove of UDG via shape and electrostatic complementarity, specific charged hydrogen bonds, and hydrophobic packing enveloping Leu-272 from a protruding UDG loop. The apparent mimicry by Ugi of DNA interactions with UDG provides both a structural mechanism for UDG binding to DNA, including the enzyme-assisted expulsion of uracil from the DNA helix, and a crystallographic basis for the design of inhibitors with scientific and therapeutic applications.
About this Structure
1UGH is a Protein complex structure of sequences from Bacillus phage pbs2 and Homo sapiens. Full crystallographic information is available from OCA.
Reference
Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA., Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA, Cell. 1995 Sep 8;82(5):701-8. PMID:7671300
Page seeded by OCA on Mon Mar 31 00:09:56 2008