We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 959
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
=== Monomere structure === | === Monomere structure === | ||
| - | Defensin-α-1 | + | Defensin-α-1 are cationic antimicrobial peptides that are synthesized in vivo as inactive precursors. <br /> |
Activation requires proteolytic excision of their anionic N-terminal inhibitory pro-peptide. The pro-peptide also specifically interacts with and inhibits the antimicrobial activity of the defensin-α-1 intermolecularly.<ref>www.ncbi.nlm.nih.gov/pmc/articles/PMC2754386/ )</ref> <br /> | Activation requires proteolytic excision of their anionic N-terminal inhibitory pro-peptide. The pro-peptide also specifically interacts with and inhibits the antimicrobial activity of the defensin-α-1 intermolecularly.<ref>www.ncbi.nlm.nih.gov/pmc/articles/PMC2754386/ )</ref> <br /> | ||
The active mature defensin-α-1 peptides consists of 29–35 amino acid residues with a molecular mass of 3–5 kDa.<br /> | The active mature defensin-α-1 peptides consists of 29–35 amino acid residues with a molecular mass of 3–5 kDa.<br /> | ||
| Line 46: | Line 46: | ||
|} | |} | ||
| - | + | There are two charged amino acid residues, <scene name='60/604478/Argglu/1'>Arg5, and Glu13</scene>, forming a conserved salt bridge, and <scene name='60/604478/Gly17/1'>Gly17</scene>, which constitutes a signature structural motif which is essential for correct folding. <br /> | |
Each defensin monomer consists of three strands of antiparallel β-sheet incorporating 60% of the residues. Two β-turns and three disulfide bonds add further restrictions to the conformational freedom of the monomer.<ref>http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2049026/</ref> | Each defensin monomer consists of three strands of antiparallel β-sheet incorporating 60% of the residues. Two β-turns and three disulfide bonds add further restrictions to the conformational freedom of the monomer.<ref>http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2049026/</ref> | ||
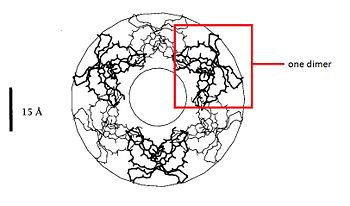
=== Dimere structure === | === Dimere structure === | ||
| - | The dimere is formed by joining identical β-strands of the two monomers together to create a symmetrical six-stranded | + | The dimere is formed by joining identical β-strands of the two monomers together to create a symmetrical six-stranded β-sheet. This extended β-sheet twists and curls to form a basket-shaped structure that has a small solvent-accessible channel passing through it. The base of the basket is hydrophobic while the top, which contains the N- and C-terminal domains of the two defensin monomers, is polar. This dimer-of-dimers may be an essential feature of defensins’interaction with membranes.<ref>Gary Fujii, Michael E.Selsted, David Eisenberg. Defensins promote fusion and lysis of negatively charged membranes. Protein Science. 1993. Cambridge University</ref> |
=== Binding with the cell membrane === | === Binding with the cell membrane === | ||
Revision as of 17:22, 4 January 2015
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Defensins-α-1
Introduction
Defensins (DEF) are a family of proteins which are involved in host defense in the epithelia of mucosal surfaces such as those of the intestin, respiratory tract, urinary tract, and vagina. They are antimicrobial and cytotoxic. All the protein of the family are distinguished by a cystein motif and are encoded on the chromozome 8.[1]
There are many defensin but in this article we'll focus on the defensin-α-1. It is a polypeptide which is found in the microbicidal granules of neutrophils. It's syntetisize in the neutrophils, which plays a role in the defense process. defensin-α-1 plays a particular role in phagocite-mediated host defense.[2]
| |||||||||||