2iy0
From Proteopedia
| Line 7: | Line 7: | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2iy0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2iy0 OCA], [http://www.ebi.ac.uk/pdbsum/2iy0 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2iy0 RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
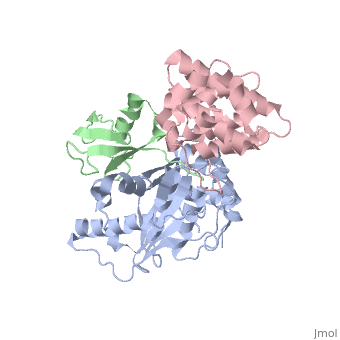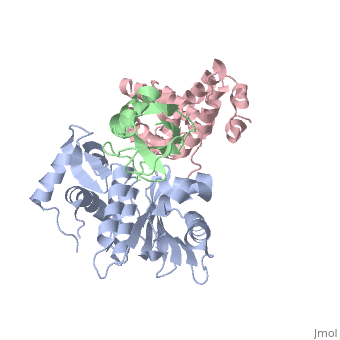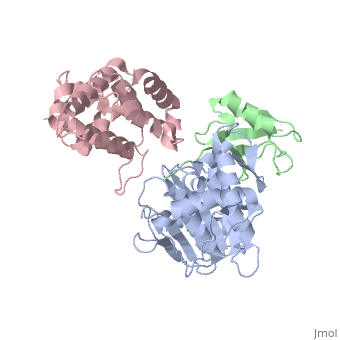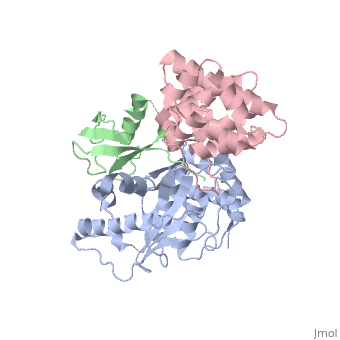
Small ubiquitin-like modifier (SUMO)-specific protease SENP1 processes SUMO-1, SUMO-2 and SUMO-3 to mature forms and deconjugates them from modified proteins. To establish the proteolytic mechanism, we determined structures of catalytically inactive SENP1 bound to SUMO-1-modified RanGAP1 and to unprocessed SUMO-1. In each case, the scissile peptide bond is kinked at a right angle to the C-terminal tail of SUMO-1 and has the cis configuration of the amide nitrogens. SENP1 preferentially processes SUMO-1 over SUMO-2, but binding thermodynamics of full-length SUMO-1 and SUMO-2 to SENP1 and K(m) values for processing are very similar. However, k(cat) values differ by 50-fold. Thus, discrimination between unprocessed SUMO-1 and SUMO-2 by SENP1 is based on a catalytic step rather than substrate binding and is likely to reflect differences in the ability of SENP1 to correctly orientate the scissile bonds in SUMO-1 and SUMO-2. | Small ubiquitin-like modifier (SUMO)-specific protease SENP1 processes SUMO-1, SUMO-2 and SUMO-3 to mature forms and deconjugates them from modified proteins. To establish the proteolytic mechanism, we determined structures of catalytically inactive SENP1 bound to SUMO-1-modified RanGAP1 and to unprocessed SUMO-1. In each case, the scissile peptide bond is kinked at a right angle to the C-terminal tail of SUMO-1 and has the cis configuration of the amide nitrogens. SENP1 preferentially processes SUMO-1 over SUMO-2, but binding thermodynamics of full-length SUMO-1 and SUMO-2 to SENP1 and K(m) values for processing are very similar. However, k(cat) values differ by 50-fold. Thus, discrimination between unprocessed SUMO-1 and SUMO-2 by SENP1 is based on a catalytic step rather than substrate binding and is likely to reflect differences in the ability of SENP1 to correctly orientate the scissile bonds in SUMO-1 and SUMO-2. | ||
| - | |||
| - | ==Disease== | ||
| - | Known diseases associated with this structure: Blood group, Cad system OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=111730 111730]], Blood group, Sd system OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=111730 111730]], Orofacial cleft 10 OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=601912 601912]] | ||
==About this Structure== | ==About this Structure== | ||
| Line 40: | Line 40: | ||
[[Category: ubl conjugation pathway]] | [[Category: ubl conjugation pathway]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:50:17 2008'' |
Revision as of 00:50, 31 March 2008
| |||||||
| , resolution 2.77Å | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
SENP1 (MUTANT) SUMO1 RANGAP
Overview
Small ubiquitin-like modifier (SUMO)-specific protease SENP1 processes SUMO-1, SUMO-2 and SUMO-3 to mature forms and deconjugates them from modified proteins. To establish the proteolytic mechanism, we determined structures of catalytically inactive SENP1 bound to SUMO-1-modified RanGAP1 and to unprocessed SUMO-1. In each case, the scissile peptide bond is kinked at a right angle to the C-terminal tail of SUMO-1 and has the cis configuration of the amide nitrogens. SENP1 preferentially processes SUMO-1 over SUMO-2, but binding thermodynamics of full-length SUMO-1 and SUMO-2 to SENP1 and K(m) values for processing are very similar. However, k(cat) values differ by 50-fold. Thus, discrimination between unprocessed SUMO-1 and SUMO-2 by SENP1 is based on a catalytic step rather than substrate binding and is likely to reflect differences in the ability of SENP1 to correctly orientate the scissile bonds in SUMO-1 and SUMO-2.
About this Structure
2IY0 is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
SUMO protease SENP1 induces isomerization of the scissile peptide bond., Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT, Nat Struct Mol Biol. 2006 Dec;13(12):1069-77. Epub 2006 Nov 12. PMID:17099698
Page seeded by OCA on Mon Mar 31 03:50:17 2008
Categories: Homo sapiens | Protein complex | Dong, C. | Naismith, J H. | Shen, L. | Gtpase activation | Hydrolase | Hydrolase/activator complex ubl conjugation | Leucine-rich repeat | Nuclear protein | Phosphorylation | Protease | Protein protein complex | Thiol protease | Ubiquitin | Ubl conjugation pathway