We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
==Insuline degrading enzyme (IDE)== | ==Insuline degrading enzyme (IDE)== | ||
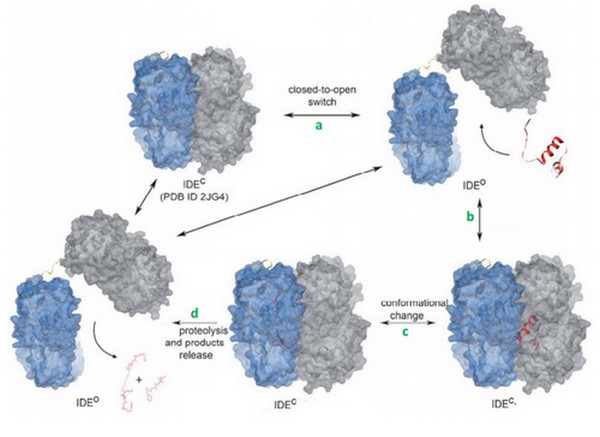
IDE (EC 3.4.24.56) is a human enzyme of the metallopeptidase family, not well-known yet. It is composed by more than 1000 residues and has a huge catalytic cavity. It is made of 2 parts linked by a loop, and it switches between an open and a close state. The size of its catalytic chamber allows the binding of peptides (70 amino acids long). IDE hydrolyzes a lot of substrates which have many differents biological activities. Its substrate can be insuline, glucagon, amyline or bradykinin. | IDE (EC 3.4.24.56) is a human enzyme of the metallopeptidase family, not well-known yet. It is composed by more than 1000 residues and has a huge catalytic cavity. It is made of 2 parts linked by a loop, and it switches between an open and a close state. The size of its catalytic chamber allows the binding of peptides (70 amino acids long). IDE hydrolyzes a lot of substrates which have many differents biological activities. Its substrate can be insuline, glucagon, amyline or bradykinin. | ||
| + | |||
| + | This enzyme hydrolyses its ubstrates by cleaving them at different points. Substrates have similar secondary structures. | ||
===Exosite: an essential element for the catalysis=== | ===Exosite: an essential element for the catalysis=== | ||
| Line 44: | Line 46: | ||
==Interactions between bradykinin and IDE== | ==Interactions between bradykinin and IDE== | ||
| - | + | Crystal structure revealed that residues 336 to 342 and 359 to 369 of IDE are involved in interactions with bradykinin. | |
| + | N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite. | ||
==Hypothetical role of bradykinin on IDE== | ==Hypothetical role of bradykinin on IDE== | ||
Revision as of 06:52, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644