We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 36: | Line 36: | ||
==Bradykinin== | ==Bradykinin== | ||
===Bradykinin biological roles=== | ===Bradykinin biological roles=== | ||
| - | Bradykinin is a short lived nonapeptide mediator of the family of kinins. It's secreted in response to an inflammatory envent and serves as a mediator of pain, inflammation and vasodilatation. | + | Bradykinin is a short lived vasoactive nonapeptide mediator of the family of kinins. It's secreted in response to an inflammatory envent and serves as a mediator of pain, inflammation and vasodilatation. |
===Bradykinin synthesis=== | ===Bradykinin synthesis=== | ||
it is secreted by the enzymatic action of kallikreins on kininogen precursors (?). Kallidin is a produt of the enzymatic action of kallikrein to kininogens. Then Kallidin is transformed into bradykinin | it is secreted by the enzymatic action of kallikreins on kininogen precursors (?). Kallidin is a produt of the enzymatic action of kallikrein to kininogens. Then Kallidin is transformed into bradykinin | ||
| Line 48: | Line 48: | ||
Crystal structure revealed that residues 336 to 342 and 359 to 369 of IDE are involved in interactions with bradykinin. | Crystal structure revealed that residues 336 to 342 and 359 to 369 of IDE are involved in interactions with bradykinin. | ||
N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite. | N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite. | ||
| + | |||
| + | The surprising thing is that bradykinin is too short to bind to both exosite and catalytic site of IDE (?). | ||
| + | |||
==Hypothetical role of bradykinin on IDE== | ==Hypothetical role of bradykinin on IDE== | ||
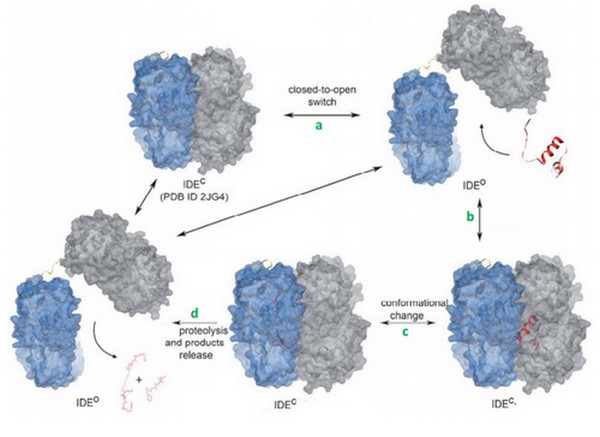
Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state. | Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state. | ||
We can also suggests that IDE binds 2 bradykinin thanks to their small lenght. | We can also suggests that IDE binds 2 bradykinin thanks to their small lenght. | ||
| + | Binding of bradykinin or other short peptides to the exosite could play a regulatory role in substrate binding and cleavage by IDE. | ||
| + | Bradykinin is supposed to reduce the size of the ctalytic chamber of IDE, and this may enhance the substrate binding and cleavage by reducing the entropy of short peptides in the chamber. Bradykinin is shown to be an activator of IDE. | ||
| + | In the other hand, it could reduce the cleavage of other substrates by interfering with their binding. | ||
| - | + | The degradation of kinins and specially bradykinin is not well understand yet. The cleavage site of bradykinin by IDE, the detailed kinetic analysis and the structural basis for recognition have to be more studied. | |
| Line 61: | Line 67: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | <ref>PMID: 2652632</ref> | ||
Revision as of 07:59, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644