We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 951
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
==Biological context== | ==Biological context== | ||
| - | Firefly, also named Photinus pyralis, is a bioluminescent insect. They are able to produce light by | + | Firefly, also named Photinus pyralis, is a bioluminescent insect. They are able to produce light by an energetic process, in order to attract its mate. The enzyme responsible of this light producing is luciferase, also known as luciferin-4-monooxygenase (EC: 1.13.12.7). It acts also as a ligase. This enzyme has various applications in the biotechnology field. In fact, it is used in chemical biology and drug trials. For example, they help to detect protein-protein interactions, to track cells in vivo in order to analysis the development of disease at the molecular level in real-time , to monitor the transcriptional and post-transcriptional regulation of specific gens, to control apoptosis, to label cancer cells, to detect environmental contaminations … |
| + | As there is no light production by mammals, luciferase is really a great tool for researchers. But this light emission depends on the environmental conditions. | ||
| + | |||
Thus, this oxydo-reductase is involved in severals reactions. | Thus, this oxydo-reductase is involved in severals reactions. | ||
=====Light emission===== | =====Light emission===== | ||
| Line 38: | Line 40: | ||
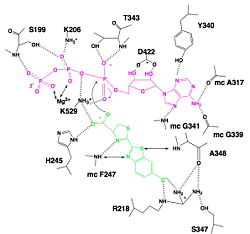
We find a signal motif in luciferase which is <scene name='60/604470/Atp_binding_signal_motif/1'>[STG]-[STG]-G-[ST]-[ST]-[TSE]-[GS]-x-[PALIVM]-K</scene> where some residue like lysine are always conserved. This pattern enables ATP binding thanks to hydrogen bonds between residues and phosphates of ATP. There is another pattern : <scene name='60/604470/Adenosine_ring_binding/1'>[YFW]-[GASW]-x-[TSA]-E</scene> which takes a particular conformation because of hydrogen bonds between residues and maintain the adenosin ring of ATP.<ref name =''fifth''>PMID:8805533</ref>, <ref>[http://www.photobiology.info/ Photobiology]</ref> | We find a signal motif in luciferase which is <scene name='60/604470/Atp_binding_signal_motif/1'>[STG]-[STG]-G-[ST]-[ST]-[TSE]-[GS]-x-[PALIVM]-K</scene> where some residue like lysine are always conserved. This pattern enables ATP binding thanks to hydrogen bonds between residues and phosphates of ATP. There is another pattern : <scene name='60/604470/Adenosine_ring_binding/1'>[YFW]-[GASW]-x-[TSA]-E</scene> which takes a particular conformation because of hydrogen bonds between residues and maintain the adenosin ring of ATP.<ref name =''fifth''>PMID:8805533</ref>, <ref>[http://www.photobiology.info/ Photobiology]</ref> | ||
=====Interaction with luciferin===== | =====Interaction with luciferin===== | ||
| - | Luciferase holds the luciferin with the specific residues <scene name='60/604470/Residues_helding_luciferin/1'>arginin 218, phenylalanin 247, serin 347 and adenin 348</scene>, still with hydrogen bounds. This bindings makes the carboxylate oxygen of luciferin points toward the α phosphate of ATP, so the oxygen is well-positionned to attack the α phosphate. This promotes the luciferin-AMP formation.<ref>PMID:8805533</ref>, <ref>[http://www.photobiology.info/ Photobiology]</ref> | + | Luciferase holds the luciferin with the specific residues <scene name='60/604470/Residues_helding_luciferin/1'>arginin 218, phenylalanin 247, serin 347 and adenin 348</scene>, still with hydrogen bounds. This bindings makes the carboxylate oxygen of luciferin points toward the α phosphate of ATP, so the oxygen is well-positionned to attack the α phosphate. This promotes the luciferin-AMP formation.<ref name =''sixth''>PMID:8805533</ref>, <ref>[http://www.photobiology.info/ Photobiology]</ref> |
=====Interaction with fatty acids===== | =====Interaction with fatty acids===== | ||
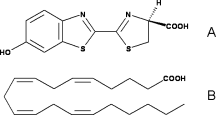
Fatty acids are highly similar to luciferin. Therefore, luciferase can use the luciferin binding site to bind fatty acids. That is why they can be used as substrates by luciferase and then, very high similar reaction as for luciferin occurs. | Fatty acids are highly similar to luciferin. Therefore, luciferase can use the luciferin binding site to bind fatty acids. That is why they can be used as substrates by luciferase and then, very high similar reaction as for luciferin occurs. | ||
Revision as of 11:35, 9 January 2015
| |||||||||||
References
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Photobiology
- ↑ Photobiology
- ↑ Photobiology
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ Hosseinkhani S. Molecular enigma of multicolor bioluminescence of firefly luciferase. Cell Mol Life Sci. 2011 Apr;68(7):1167-82. doi: 10.1007/s00018-010-0607-0. Epub, 2010 Dec 28. PMID:21188462 doi:http://dx.doi.org/10.1007/s00018-010-0607-0
- ↑ Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell Mol Life Sci. 2010 Feb;67(3):387-404. Epub 2009 Oct 27. PMID:19859663 doi:10.1007/s00018-009-0170-8