We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | <ref>doi: 10.1021/bi801192h</ref> | ||
==Context== | ==Context== | ||
| Line 37: | Line 36: | ||
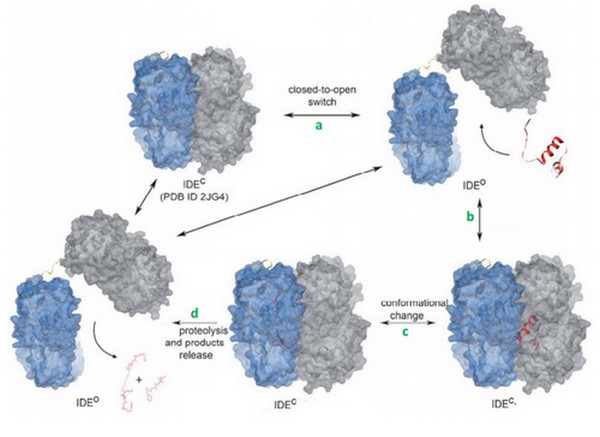
IDE can bind many substrates to its exosite and must recognize them. In order to accomplish this task, IDE uses many tools. | IDE can bind many substrates to its exosite and must recognize them. In order to accomplish this task, IDE uses many tools. | ||
First, the lenght of the substrate peptide is essential. The peptide must be long enough to touch both the exosite and the catalytic site. Second, the peptide must be short enough in order to enter the catalytic chamber. In the case of the bradykinin, the length is too small to touch the catalytic site when it binds the exosite. | First, the lenght of the substrate peptide is essential. The peptide must be long enough to touch both the exosite and the catalytic site. Second, the peptide must be short enough in order to enter the catalytic chamber. In the case of the bradykinin, the length is too small to touch the catalytic site when it binds the exosite. | ||
| - | Tang et al have shown that bradykinin is too small to bind both at the exosite and the catalytic side. They prooved that IDE binds 2 bradykinin at the same time : the first one interacts with the exosite and the second one touches the catalytic site. The bradykinin in the catalytic site is the one which is going to be cleaved by the enzyme. | + | Tang et al(??) have shown that bradykinin is too small to bind both at the exosite and the catalytic side. They prooved that IDE binds 2 bradykinin at the same time : the first one interacts with the exosite and the second one touches the catalytic site. The bradykinin in the catalytic site is the one which is going to be cleaved by the enzyme. |
This can be a discriminate factor, because IDE binds <scene name='60/604491/Bradykinin/1'>2 bradykinins</scene> at the same time. | This can be a discriminate factor, because IDE binds <scene name='60/604491/Bradykinin/1'>2 bradykinins</scene> at the same time. | ||
| Line 50: | Line 49: | ||
Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state. | Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state. | ||
We can also suggests that IDE binds <scene name='60/604491/Bradykinin/1'>2 bradykinins</scene> thanks to their small lenght. | We can also suggests that IDE binds <scene name='60/604491/Bradykinin/1'>2 bradykinins</scene> thanks to their small lenght. | ||
| - | Binding of bradykinin or other short peptides to the exosite could play a regulatory role in substrate binding and cleavage by IDE. | + | Binding of bradykinin or other short peptides to the exosite could play a regulatory role in substrate binding and cleavage by IDE<ref>doi: 10.1021/bi801192h.</ref>. |
Bradykinin is supposed to reduce the size of the ctalytic chamber of IDE, and this may enhance the substrate binding and cleavage by reducing the entropy of short peptides in the chamber. Bradykinin is shown to be an activator of IDE. | Bradykinin is supposed to reduce the size of the ctalytic chamber of IDE, and this may enhance the substrate binding and cleavage by reducing the entropy of short peptides in the chamber. Bradykinin is shown to be an activator of IDE. | ||
In the other hand, it could reduce the cleavage of other substrates by interfering with their binding. | In the other hand, it could reduce the cleavage of other substrates by interfering with their binding. | ||
Revision as of 20:50, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ doi: https://dx.doi.org/10.1021/bi801192h.