We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 38: | Line 38: | ||
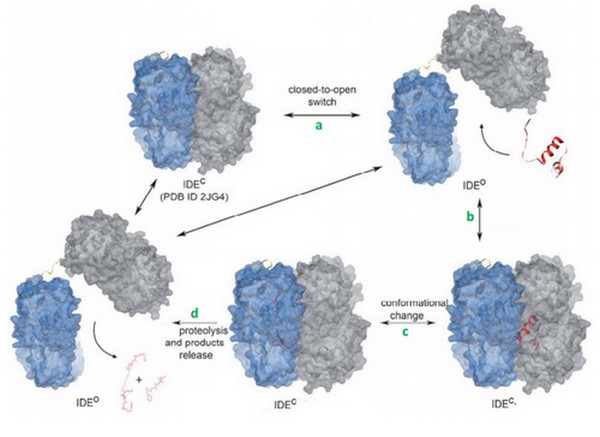
IDE catalytic site has a high affinity for hydrophobic and basic. Bradykinin is essentially composed by proline and arginine, which are basic amino acids. So, bradykinin structure may explain this strange interaction. | IDE catalytic site has a high affinity for hydrophobic and basic. Bradykinin is essentially composed by proline and arginine, which are basic amino acids. So, bradykinin structure may explain this strange interaction. | ||
| - | |||
| - | |||
Crystal structure revealed that <scene name='60/604491/Residues_336_to_342/1'>residues 336 to 342</scene> and <scene name='60/604491/Residues_359_to_369/1'>residues 359 to 369</scene> of IDE are involved in interactions with bradykinin. | Crystal structure revealed that <scene name='60/604491/Residues_336_to_342/1'>residues 336 to 342</scene> and <scene name='60/604491/Residues_359_to_369/1'>residues 359 to 369</scene> of IDE are involved in interactions with bradykinin. | ||
N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite. | N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite. | ||
| + | |||
| + | Im et al<ref>doi: 10.1074/jbc.M701590200</ref> suggested that ATP increases the activtity of IDE with its small substrates like braydkinin. | ||
==Hypothetical role of bradykinin on IDE== | ==Hypothetical role of bradykinin on IDE== | ||
Revision as of 21:31, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Song ES, Juliano MA, Juliano L, Hersh LB. Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. J Biol Chem. 2003 Dec 12;278(50):49789-94. Epub 2003 Oct 2. PMID:14527953 doi:http://dx.doi.org/10.1074/jbc.M308983200
- ↑ Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007 Aug 31;282(35):25453-63. Epub 2007 Jul 5. PMID:17613531 doi:10.1074/jbc.M701590200
- ↑ Malito E, Ralat LA, Manolopoulou M, Tsay JL, Wadlington NL, Tang WJ. Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme. Biochemistry. 2008 Nov 6. PMID:18986166 doi:10.1021/bi801192h