Sandbox Reserved 966
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
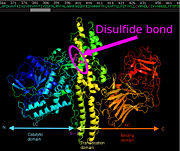
[[Image:BoNT.jpg|thumb|'''Figure 2 :''' Global aspect of C.botulinum neurotoxin serotype A (made using PyMol and modified with Inkscape by Xavier Hartmann) ]] | [[Image:BoNT.jpg|thumb|'''Figure 2 :''' Global aspect of C.botulinum neurotoxin serotype A (made using PyMol and modified with Inkscape by Xavier Hartmann) ]] | ||
The neurotoxin inactivates neurotransmitter release owing to its metalloprotease activity. It targets the presynaptic membrane of peripheral nerve terminals using a binding mode based on the use of two independent receptors: a polysialoganglioside (PSG) and a protein receptor in the lumen of synaptic vesicles<ref>PMID: 23239349</ref>. ''Clostridium botulinum'' neurotoxin is initially synthesized as a single polypeptide chain of ≈150 kDa and is then cleaved by a protease to yield the mature toxin, which consists of a light chain (LC which is 50 kDa) and a heavy chain (HC which is 100 kDa). The heavy chain presents the PSG binding domain and is therefore implied in the entry of the neurotoxin in the nerve cell thanks to vesicles; then the disulfide bond is broken and the light chain is released. | The neurotoxin inactivates neurotransmitter release owing to its metalloprotease activity. It targets the presynaptic membrane of peripheral nerve terminals using a binding mode based on the use of two independent receptors: a polysialoganglioside (PSG) and a protein receptor in the lumen of synaptic vesicles<ref>PMID: 23239349</ref>. ''Clostridium botulinum'' neurotoxin is initially synthesized as a single polypeptide chain of ≈150 kDa and is then cleaved by a protease to yield the mature toxin, which consists of a light chain (LC which is 50 kDa) and a heavy chain (HC which is 100 kDa). The heavy chain presents the PSG binding domain and is therefore implied in the entry of the neurotoxin in the nerve cell thanks to vesicles; then the disulfide bond is broken and the light chain is released. | ||
| - | It is the light chain that carries the metalloprotease activity. Neurotoxin serotype A cleaves a peptide bond of a protein belonging to the SNARE family [http://en.wikipedia.org/wiki/SNARE_(protein) ''Wikipedia page''] (involved in the phenomenon of vesicle fusion) and especially the Gln197 - Arg198 peptide bond of SNAP-25 [http://en.wikipedia.org/wiki/SNAP25 ''Wikipedia page''] blocking the release of neurotransmitter release inducing paralysis. The BoNT LCs are remarkable among proteases for the extremely long substrates required for efficient proteolysis. With a minimum substrate of 17 amino acids (SNKTRIDQAN[[QR]]ATKML, cleavage site in red), the BoNT/A protease accepts shorter peptide substrates than any of the other serotypes. In comparison, other microbial metalloproteases have been found to have activity against substrates as short as dipeptides.<ref>PMID: 18457419</ref> Structural and biochemical data on the BoNT/ A-LC suggest that most of the specific interactions between the enzyme and the SNAP-25 substrate occur at sites remote from the active site. Indeed, the only amino acid within the cleavage sequence that is required for efficient proteolysis is the P1′ Arg198 <ref>PMID:15592454</ref>. | + | |
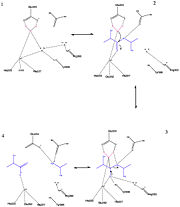
| + | It is the light chain that carries the metalloprotease activity. Neurotoxin serotype A cleaves a peptide bond of a protein belonging to the SNARE family [http://en.wikipedia.org/wiki/SNARE_(protein) ''Wikipedia page''] (involved in the phenomenon of vesicle fusion) and especially the Gln197 - Arg198 peptide bond of SNAP-25 [http://en.wikipedia.org/wiki/SNAP25 ''Wikipedia page''] blocking the release of neurotransmitter release, inducing paralysis. The BoNT LCs are remarkable among proteases for the extremely long substrates required for efficient proteolysis. With a minimum substrate of 17 amino acids (SNKTRIDQAN[[QR]]ATKML, cleavage site in red), the BoNT/A protease accepts shorter peptide substrates than any of the other serotypes. In comparison, other microbial metalloproteases have been found to have activity against substrates as short as dipeptides.<ref>PMID: 18457419</ref> Structural and biochemical data on the BoNT/ A-LC suggest that most of the specific interactions between the enzyme and the SNAP-25 substrate occur at sites remote from the active site. Indeed, the only amino acid within the cleavage sequence that is required for efficient proteolysis is the P1′ Arg198 <ref>PMID:15592454</ref>. | ||
[[Image:Mecanismefinal.jpg|thumb|'''Figure 3 :''' Enzymatic mechanism of BoNT/A. Steps are numbered (made using ChemDraw Ultra by Xavier Hartmann) ]] | [[Image:Mecanismefinal.jpg|thumb|'''Figure 3 :''' Enzymatic mechanism of BoNT/A. Steps are numbered (made using ChemDraw Ultra by Xavier Hartmann) ]] | ||
| Line 58: | Line 59: | ||
This pathologic can be treated by the same methode. Every anomalous behaviour of muscle tissue can be fixed thanks to the paralyzing effect of the toxin. It is based on the muscles relaxation. | This pathologic can be treated by the same methode. Every anomalous behaviour of muscle tissue can be fixed thanks to the paralyzing effect of the toxin. It is based on the muscles relaxation. | ||
=== Cervical dystonia === | === Cervical dystonia === | ||
| - | BoNT/A is used against cervical dystonia. But it can become inefficient after a | + | BoNT/A is used against cervical dystonia. But it can become inefficient after a period of use. |
Revision as of 02:27, 10 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Structure of the "Clostridium botulinum" neurotoxin serotype A light chain with Zn2+ cofactor bound
| |||||||||||