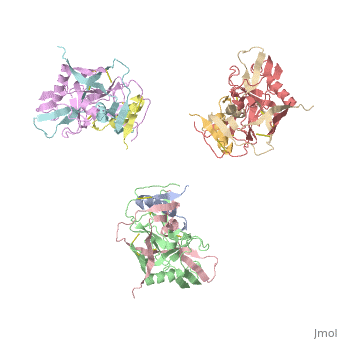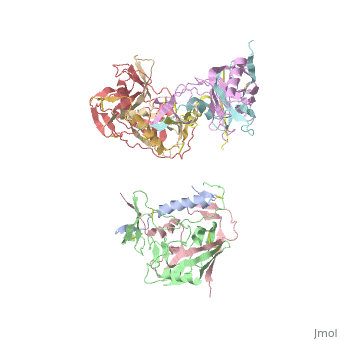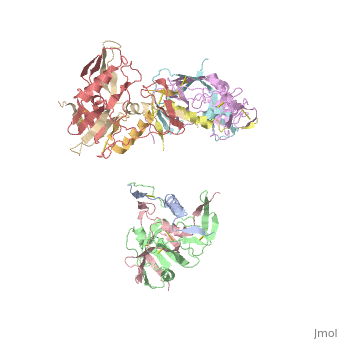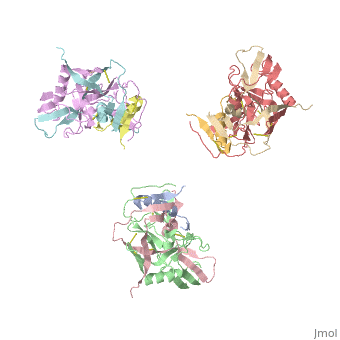We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hiv env proteins
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Env Structure == | == Env Structure == | ||
The viral Env protein, as mentioned above, is composed of gp120 and gp41 that forms trimers on the viral membrane surface. | The viral Env protein, as mentioned above, is composed of gp120 and gp41 that forms trimers on the viral membrane surface. | ||
| - | [[Image:Trimeric_gp_spike_on_HIV1.png|thumb| | + | [[Image:Trimeric_gp_spike_on_HIV1.png|left|thumb|400px|<ref name=Nick>PMID: 18668044 </ref>]] |
{{Clear}} | {{Clear}} | ||
The image to the right, displays the undecorated Env trimer. Within the gp120 monomeric proteins are variable loops that undergo conformational changes that support binding or restrict binding of receptors such as CD4 or antibodies. Two important loops in this regard are the V1/V2 loops. The V1/V2 loops are located at the apex of the structure in the center of the three monomeric gp120 proteins. Depending on the gp120 conformation changes, the three gp41 transmembrane proteins, located beneath the gp120 surface proteins (located by the white arrow in F2.b), will undergo "irreversible refolding," bringing the viral membrane and host cell membrane closer together, consequently inducing fusion <ref name=Guan>PMID: 21076402 </ref>. | The image to the right, displays the undecorated Env trimer. Within the gp120 monomeric proteins are variable loops that undergo conformational changes that support binding or restrict binding of receptors such as CD4 or antibodies. Two important loops in this regard are the V1/V2 loops. The V1/V2 loops are located at the apex of the structure in the center of the three monomeric gp120 proteins. Depending on the gp120 conformation changes, the three gp41 transmembrane proteins, located beneath the gp120 surface proteins (located by the white arrow in F2.b), will undergo "irreversible refolding," bringing the viral membrane and host cell membrane closer together, consequently inducing fusion <ref name=Guan>PMID: 21076402 </ref>. | ||
| Line 16: | Line 16: | ||
== gp120 Binding == | == gp120 Binding == | ||
gp120 is responsible for the binding of HIV to CD4 cells. "Binding of gp120 to the primary receptor CD4 and coreceptor (for example, CCR5 and CXCR4) induces large conformational changes, which then trigger dissociation of gp120 and a cascade of refolding events in gp41" <ref name=Guan>PMID: 21076402 </ref>. In the unliganded state, the <scene name='Sandbox/Gp120_v1_v2_loops/1'>V1/V2 loops</scene> are located at the center of the apex of the trimer and hold the trimer together. However, when <scene name='Sandbox/Gp120_v1_v2_loops/2'>CD4</scene> binds to gp120, the V1/V2 loops undergo an outward rotational change that expose the central gp41 proteins at the base of the Env protein. gp120 is often complexed with <scene name='Hiv_env_proteins/Cd4_gp120_complex/1'>CD4</scene> and an antibody Fab (17b-Fab). "The 17b antibody has been shown to stabilize and lock gp120 in the CD4-bound conformation" <ref name=Nick>PMID: 18668044 </ref>. "Direct interatomic contacts are made between 22 CD4 residues and 26 gp120 amino-acid residues. These include 219 van der Waals contacts and 12 hydrogen bonds" <ref name=Nich>PMID: 9641677 </ref>. "Residues in contact are concentrated in the <scene name='Hiv_env_proteins/Cd4_gp120_interface/1'>span</scene> from 25 to 64 of CD4, but they are distributed over six segments of gp120" <ref name=Nich>PMID: 9641677 </ref>. However, the most important gp120/CD4 <scene name='Hiv_env_proteins/Cd4_gp120_complex/2'>interactions</scene> are between "Phe 43 and Arg 59 of CD4 [which] make multiple contacts centered on residues Asp 368, Glu 370 and Trp427 of gp120, which are all conserved among primate immunodeficiency viruses" <ref name=Nich>PMID: 9641677 </ref>. | gp120 is responsible for the binding of HIV to CD4 cells. "Binding of gp120 to the primary receptor CD4 and coreceptor (for example, CCR5 and CXCR4) induces large conformational changes, which then trigger dissociation of gp120 and a cascade of refolding events in gp41" <ref name=Guan>PMID: 21076402 </ref>. In the unliganded state, the <scene name='Sandbox/Gp120_v1_v2_loops/1'>V1/V2 loops</scene> are located at the center of the apex of the trimer and hold the trimer together. However, when <scene name='Sandbox/Gp120_v1_v2_loops/2'>CD4</scene> binds to gp120, the V1/V2 loops undergo an outward rotational change that expose the central gp41 proteins at the base of the Env protein. gp120 is often complexed with <scene name='Hiv_env_proteins/Cd4_gp120_complex/1'>CD4</scene> and an antibody Fab (17b-Fab). "The 17b antibody has been shown to stabilize and lock gp120 in the CD4-bound conformation" <ref name=Nick>PMID: 18668044 </ref>. "Direct interatomic contacts are made between 22 CD4 residues and 26 gp120 amino-acid residues. These include 219 van der Waals contacts and 12 hydrogen bonds" <ref name=Nich>PMID: 9641677 </ref>. "Residues in contact are concentrated in the <scene name='Hiv_env_proteins/Cd4_gp120_interface/1'>span</scene> from 25 to 64 of CD4, but they are distributed over six segments of gp120" <ref name=Nich>PMID: 9641677 </ref>. However, the most important gp120/CD4 <scene name='Hiv_env_proteins/Cd4_gp120_complex/2'>interactions</scene> are between "Phe 43 and Arg 59 of CD4 [which] make multiple contacts centered on residues Asp 368, Glu 370 and Trp427 of gp120, which are all conserved among primate immunodeficiency viruses" <ref name=Nich>PMID: 9641677 </ref>. | ||
| - | [[Image:3D structure of the HIV-1 spike in complex with CD4 and 17b- Fab.png|thumb| | + | [[Image:3D structure of the HIV-1 spike in complex with CD4 and 17b- Fab.png|left|thumb|400px|<ref name=Nick>PMID: 18668044 </ref>]] |
{{Clear}} | {{Clear}} | ||
| Line 31: | Line 31: | ||
== gp41 == | == gp41 == | ||
gp41 is a transmembrane protein on the viral envelope of HIV, with its "C-terminal transmembrane segment inserted in the viral membrane" and its N-terminal outside the viral membrane beneath the gp120 proteins <ref name=Guan>PMID: 21076402 </ref>. gp41 facilitates fusion of HIV with the host cell. There are 3 different conformations that gp41 can exist in: prefusion state, prehairpin intermediate, and post fusion state. In the prefusion state gp41 exists as the transmembrane segment mentioned above. Upon binding of gp120 to CD4, gp41 enters the prehairpin intermediate, where it extends into the host cell and the C-terminal segment remains in the viral envelope. Further rearrangement induces the post fusion state, where gp41 is completely enters the host cell, causing fusion of the membranes. | gp41 is a transmembrane protein on the viral envelope of HIV, with its "C-terminal transmembrane segment inserted in the viral membrane" and its N-terminal outside the viral membrane beneath the gp120 proteins <ref name=Guan>PMID: 21076402 </ref>. gp41 facilitates fusion of HIV with the host cell. There are 3 different conformations that gp41 can exist in: prefusion state, prehairpin intermediate, and post fusion state. In the prefusion state gp41 exists as the transmembrane segment mentioned above. Upon binding of gp120 to CD4, gp41 enters the prehairpin intermediate, where it extends into the host cell and the C-terminal segment remains in the viral envelope. Further rearrangement induces the post fusion state, where gp41 is completely enters the host cell, causing fusion of the membranes. | ||
| - | [[Image:Structure complex gp41-post 1281Fab.png|thumb| | + | [[Image:Structure complex gp41-post 1281Fab.png|left|thumb|400px|<ref name=Guan>PMID: 21076402 </ref>]] |
{{Clear}} | {{Clear}} | ||
There are three main types of antibodies that can potentially bind gp 41: MPER antibodies, cluter I antibodies, and cluster II antibodies. These are either neutralizing antibodies or non-neutralizing antibodies. MPER "neutralizing antibodies target a region on gp41 adjacent to the viral membrane, called the <scene name='Hiv_env_proteins/Gp41_mper/1'>membrane-proximal external region</scene> (MPER)" represented by the red region <ref name=Guan>PMID: 21076402 </ref>. Two broadly neutralizing proteins that target this region in the prehairpin-intermediate state are: 4E10 and 2F5. However, non-neutralizing antibodies are more prevalent in infected individuals. Cluster I antibodies target a part of gp41 that is not readily accessible while "Cluster II antibodies recognize another <scene name='Hiv_env_proteins/Gp41_mper/2'>immunodominant segment</scene> (residues 644–663) next to the MPER" <ref name=Guan>PMID: 21076402 </ref>. Cluster II antibodies are non-neutralizing because they bind to gp41 in the <scene name='Hiv_env_proteins/Gp41_181_interaction/1'>post fusion</scene> conformation, after the fusion is already complete or when gp120 proteins are prematurely shed. Cluster II epitopes, like the six alpha helix bundle, facilitate HIV-1 immune evasion by triggering the production cluster II antibodies that are non-neutralizing. "HIV-1 may thereby exploit the envelope stability as one of its immune-evasion tactics to distract the immune system from the native, functional trimers" <ref name=Guan>PMID: 21076402 </ref>. "The stable postfusion conformation of gp41 probably serves as a decoy to help HIV-1 evade the immune system and induce ineffective antibody responses in infected people. It has been shown that unique B-cell clones targeting cluster II epitopes account for 49% of all anti-gp41-reactive B cells" <ref name=Guan>PMID: 21076402 </ref>. | There are three main types of antibodies that can potentially bind gp 41: MPER antibodies, cluter I antibodies, and cluster II antibodies. These are either neutralizing antibodies or non-neutralizing antibodies. MPER "neutralizing antibodies target a region on gp41 adjacent to the viral membrane, called the <scene name='Hiv_env_proteins/Gp41_mper/1'>membrane-proximal external region</scene> (MPER)" represented by the red region <ref name=Guan>PMID: 21076402 </ref>. Two broadly neutralizing proteins that target this region in the prehairpin-intermediate state are: 4E10 and 2F5. However, non-neutralizing antibodies are more prevalent in infected individuals. Cluster I antibodies target a part of gp41 that is not readily accessible while "Cluster II antibodies recognize another <scene name='Hiv_env_proteins/Gp41_mper/2'>immunodominant segment</scene> (residues 644–663) next to the MPER" <ref name=Guan>PMID: 21076402 </ref>. Cluster II antibodies are non-neutralizing because they bind to gp41 in the <scene name='Hiv_env_proteins/Gp41_181_interaction/1'>post fusion</scene> conformation, after the fusion is already complete or when gp120 proteins are prematurely shed. Cluster II epitopes, like the six alpha helix bundle, facilitate HIV-1 immune evasion by triggering the production cluster II antibodies that are non-neutralizing. "HIV-1 may thereby exploit the envelope stability as one of its immune-evasion tactics to distract the immune system from the native, functional trimers" <ref name=Guan>PMID: 21076402 </ref>. "The stable postfusion conformation of gp41 probably serves as a decoy to help HIV-1 evade the immune system and induce ineffective antibody responses in infected people. It has been shown that unique B-cell clones targeting cluster II epitopes account for 49% of all anti-gp41-reactive B cells" <ref name=Guan>PMID: 21076402 </ref>. | ||
Revision as of 12:50, 15 January 2015
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Frey G, Chen J, Rits-Volloch S, Freeman MM, Zolla-Pazner S, Chen B. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat Struct Mol Biol. 2010 Dec;17(12):1486-91. Epub 2010 Nov 14. PMID:21076402 doi:10.1038/nsmb.1950
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008 Sep 4;455(7209):109-13. Epub 2008 Jul 30. PMID:18668044 doi:10.1038/nature07159
- ↑ 3.0 3.1 3.2 Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998 Jun 18;393(6686):648-59. PMID:9641677 doi:10.1038/31405
- ↑ 4.0 4.1 4.2 4.3 Chen L, Do Kwon Y, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009 Nov 20;326(5956):1123-7. PMID:19965434 doi:326/5956/1123
- ↑ Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Karim QA, Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012 Nov;18(11):1688-92. doi: 10.1038/nm.2985. Epub 2012 Oct 21. PMID:23086475 doi:10.1038/nm.2985