We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
RiAFP
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
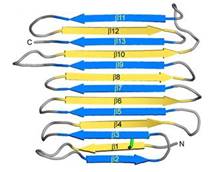
[[Image:Fig2_B.jpg|frame|alt=Puzzle globe|Fig. 1. Secondary structure diagram]] The crystallographic structure of RiAFP was defined recently<ref>DOI 10.1074/jbc.M113.450973</ref>. RiAFP has a novel β-solenoid architecture that forms <scene name='60/607864/Beta_sheets/1'>β-sandwich</scene>. This sandwich is composed of two parallel remarkably regular <scene name='60/607864/Beta_sheets_colored/1'> 6 and 7 stranded-sheets</scene>. The β-sheets lie on top of each other with the upper and lower strands parallel but in the opposite orientation. β1 and β2 strands are untiparallel to the other strands in each sandwich plane. Two ends deviate from β helix regularity by forming <scene name='60/607864/Beta_sheets_capping/2'>capping structures</scene> (see Figure 1). These capping structures help to prevent end-to-end associations that would spoil the solubility of RiAFP and lead to oligomerization and aggregation. | [[Image:Fig2_B.jpg|frame|alt=Puzzle globe|Fig. 1. Secondary structure diagram]] The crystallographic structure of RiAFP was defined recently<ref>DOI 10.1074/jbc.M113.450973</ref>. RiAFP has a novel β-solenoid architecture that forms <scene name='60/607864/Beta_sheets/1'>β-sandwich</scene>. This sandwich is composed of two parallel remarkably regular <scene name='60/607864/Beta_sheets_colored/1'> 6 and 7 stranded-sheets</scene>. The β-sheets lie on top of each other with the upper and lower strands parallel but in the opposite orientation. β1 and β2 strands are untiparallel to the other strands in each sandwich plane. Two ends deviate from β helix regularity by forming <scene name='60/607864/Beta_sheets_capping/2'>capping structures</scene> (see Figure 1). These capping structures help to prevent end-to-end associations that would spoil the solubility of RiAFP and lead to oligomerization and aggregation. | ||
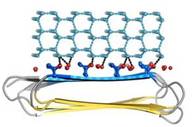
The three residues in β-strand 11 at the C terminus <scene name='60/607864/Gln110_gln112_ile114/1'>(Gln110, Gln112, and Ile114)</scene> that are too bulky to be accommodated into the core may also contribute to the capping structure to prevent amyloid-like polymerization. RiAFP solenoid possesses compressed nature with average distance between the sheets of only 6 Å. In the core of RiAFP, the side chains within apposed β-strands from the two β-sheets are staggered, allowing the side chains to interdigitate and pack tightly against one another. Most of the <scene name='60/607864/Core_structure/1'>side chains</scene> in the core are from <span style="color:pink;background-color:black;font-weight:bold;">Ala</span>, <span style="color:green;background-color:black;font-weight:bold;">Ser</span> and <span style="color:yellow;background-color:black;font-weight:bold;">Thr</span>. Those residues create a more compact fold that may contribute to the high stability and antifreeze activity of RiAFP. Within the core there are <scene name='60/607864/Hydrogen_bonds/2'>hydrogen bonds</scene> between Thr-Ser (65-55, 85-75, 132-124 respectively) and one <scene name='60/607864/S-s_bond/1'>disulfide bond</scene> between Cys4-Cys21, that contributes to stabilization of the whole structure. The β-turns in the structure contain mostly Gly or Pro residues. | The three residues in β-strand 11 at the C terminus <scene name='60/607864/Gln110_gln112_ile114/1'>(Gln110, Gln112, and Ile114)</scene> that are too bulky to be accommodated into the core may also contribute to the capping structure to prevent amyloid-like polymerization. RiAFP solenoid possesses compressed nature with average distance between the sheets of only 6 Å. In the core of RiAFP, the side chains within apposed β-strands from the two β-sheets are staggered, allowing the side chains to interdigitate and pack tightly against one another. Most of the <scene name='60/607864/Core_structure/1'>side chains</scene> in the core are from <span style="color:pink;background-color:black;font-weight:bold;">Ala</span>, <span style="color:green;background-color:black;font-weight:bold;">Ser</span> and <span style="color:yellow;background-color:black;font-weight:bold;">Thr</span>. Those residues create a more compact fold that may contribute to the high stability and antifreeze activity of RiAFP. Within the core there are <scene name='60/607864/Hydrogen_bonds/2'>hydrogen bonds</scene> between Thr-Ser (65-55, 85-75, 132-124 respectively) and one <scene name='60/607864/S-s_bond/1'>disulfide bond</scene> between Cys4-Cys21, that contributes to stabilization of the whole structure. The β-turns in the structure contain mostly Gly or Pro residues. | ||
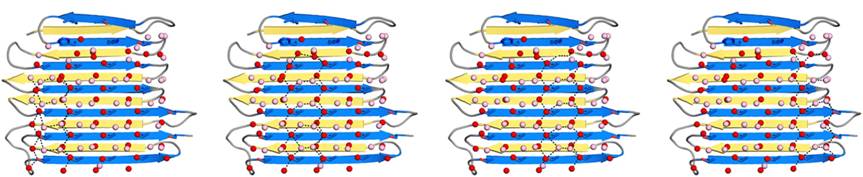
| - | The asymmetric unit comprises two RiAFP molecules juxtaposed with their ice-binding surfaces, however the protein is monomer in the solution. | + | The <scene name='60/607864/Assymetric_unit/1'>asymmetric unit</scene> comprises two RiAFP molecules juxtaposed with their ice-binding surfaces, however the protein is monomer in the solution. |
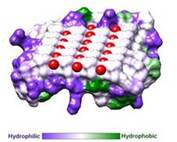
== Ice Binding Surface (IBS) == | == Ice Binding Surface (IBS) == | ||
Revision as of 23:22, 24 January 2015
| |||||||||||
3D structures of antifreeze protein
References
- ↑ Jia Z, Davies PL. Antifreeze proteins: an unusual receptor-ligand interaction. Trends Biochem Sci. 2002 Feb;27(2):101-6. PMID:11852248
- ↑ Chantelle J. Capicciotti, Malay Doshi and Robert N. Ben (2013). Ice Recrystallization Inhibitors: From Biological Antifreezes to Small Molecules, Recent Developments in the Study of Recrystallization, Prof. Peter Wilson (Ed.), ISBN: 978-953-51-0962-4, InTech doi:http://dx.doi.org/10.5772/54992
- ↑ Drori R, Celik Y, Davies PL, Braslavsky I. Ice-binding proteins that accumulate on different ice crystal planes produce distinct thermal hysteresis dynamics. J R Soc Interface. 2014 Sep 6;11(98):20140526. doi: 10.1098/rsif.2014.0526. PMID:25008081 doi:http://dx.doi.org/10.1098/rsif.2014.0526
- ↑ doi: https://dx.doi.org/10.1016/S0006-3495(91)82234-2
- ↑ Hakim A, Nguyen JB, Basu K, Zhu DF, Thakral D, Davies PL, Isaacs FJ, Modis Y, Meng W. Crystal structure of an insect antifreeze protein and its implications for ice binding. J Biol Chem. 2013 Apr 26;288(17):12295-304. doi: 10.1074/jbc.M113.450973. Epub, 2013 Mar 12. PMID:23486477 doi:10.1074/jbc.M113.450973