Human ABO(H) Blood Group Glycosyltransferases
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
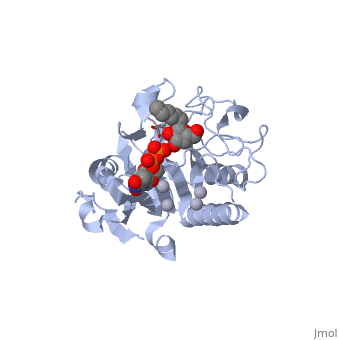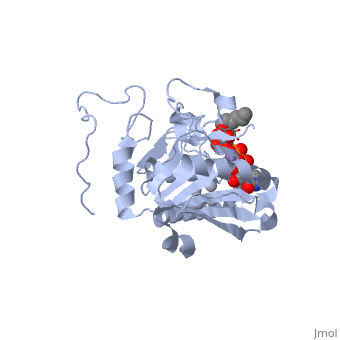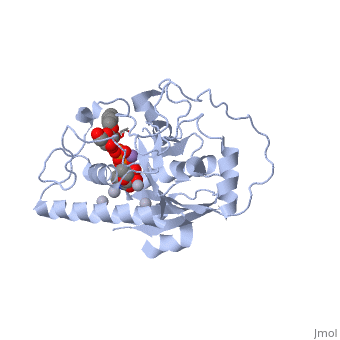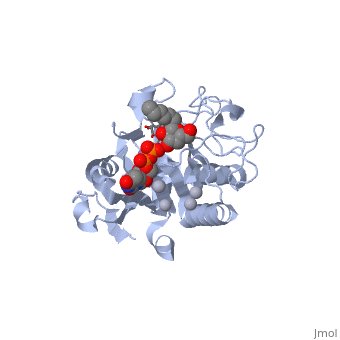
| + | '''General topology of GTA and GTB''' | ||
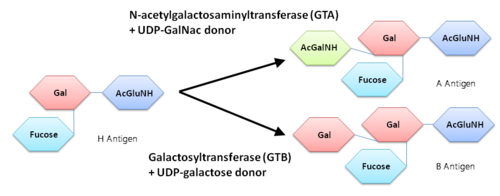
| + | Both GTs consist of 354 residues and they only differ by four "critical" amino acids. | ||
| + | the polypeptide chain is organized in two domains separated by a cleft, approximately 13 Å wide, containing the active site which consist all four critical amino acid residues. The N-terminal, includes a [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann fold] and recognizes the nucleotide donor, whereas the disaccharide acceptor binding site is formed by residues in the C-terminal domain in combination with bound UDP. | ||
| + | In the middle of the cleft located a DXD motif, highly conserved in a large number of glycosyltransferases, which coordinates the Mn2+ ion and was suggested to have a role in catalysis. | ||
| + | |||
| + | '''H-antigen and UDP substrate binding''' | ||
| + | Studies show that binding of the donor must precede acceptor binding. Indeed, the donor is a key preliminary step in the formation of the acceptor-binding site as both sugar residues of the acceptor make strong hydrogen bonds with the beta-phosphate group of the UDP. | ||
| + | The donor binds to the enzyme through Phe 121, Ile 123, Tyr 126 and Asp 213. | ||
| + | The H-antigen acceptor interacts with the enzyme primarily through the galactose O4-hydroxyl group, making hydrogen bond interactions with the side chains of His 233 and Glu 303, and the Gal O6-hydroxyl group, which hydrogen bonds to Thr 245 and the Fuc O4'-hydroxyl to Asp 326. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | + | ||
<scene name='69/691577/Bbbb_4res/1'>4 critical residuse</scene> | <scene name='69/691577/Bbbb_4res/1'>4 critical residuse</scene> | ||
Revision as of 21:29, 25 January 2015
| |||||||||||