Binding site of AChR
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
There are two kinds of acetylcholine receptor in nature: [http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor nicotinic acetylcholine receptors] and [http://en.wikipedia.org/wiki/Muscarinic_acetylcholine_receptor muscarinic acetylcholine receptors]. The nicotinic acetylcholine receptor(nAChR) is a pentameric ligand-gated ion channel activated by binding of acetylcholine in nature. In this page we will show the structure of binding site of nAChR by a complex between α-bungarotoxin and a mimotope peptide. | There are two kinds of acetylcholine receptor in nature: [http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor nicotinic acetylcholine receptors] and [http://en.wikipedia.org/wiki/Muscarinic_acetylcholine_receptor muscarinic acetylcholine receptors]. The nicotinic acetylcholine receptor(nAChR) is a pentameric ligand-gated ion channel activated by binding of acetylcholine in nature. In this page we will show the structure of binding site of nAChR by a complex between α-bungarotoxin and a mimotope peptide. | ||
| - | == Basic structure of AChR == | + | == Basic structure and opening mechanism of AChR == |
| - | Pentameric ligand gated ion channels (pLGIC), or [http://en.wikipedia.org/wiki/Cys-loop_receptors Cys-loop receptors],are a group of transmembrane ion channel proteins which open to allow ions such as Na+, K+, Ca2+, or Cl- to pass through the membrane in response to the binding of a chemical messenger, such as a neurotransmitter<ref> Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.</ref>. In overall organization, the <scene name='68/688431/Plgics/1'>pLGICs</scene> have five subunits. The five subunits are arranged in a barrel-like manner around a central symmetry axis that coincides with the ion permeation pathway.<ref>PMID:24167270</ref> In each subunit, the extracellular domin(ECD) of pLGIC encompasses 10β-strands that are organized as a sandwich of two tightly interacting β-sheets, while the transmembrane domain(TMD) folds into a bundle of four α-helices (M1, M2, M3, M4). | + | Acetylcholine receptor is a member of pentameric ligand gated ion channels family,which share the similar structure. Pentameric ligand gated ion channels (pLGIC), or [http://en.wikipedia.org/wiki/Cys-loop_receptors Cys-loop receptors],are a group of transmembrane ion channel proteins which open to allow ions such as Na+, K+, Ca2+, or Cl- to pass through the membrane in response to the binding of a chemical messenger, such as a neurotransmitter<ref> Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.</ref>. In overall organization, the <scene name='68/688431/Plgics/1'>pLGICs</scene> have five subunits. The five subunits are arranged in a barrel-like manner around a central symmetry axis that coincides with the ion permeation pathway.<ref>PMID:24167270</ref> In each subunit, the extracellular domin(ECD) of pLGIC encompasses 10β-strands that are organized as a sandwich of two tightly interacting β-sheets, while the transmembrane domain(TMD) folds into a bundle of four α-helices (M1, M2, M3, M4). |
| - | X-ray structure of homologues of the extracellular domain(ECD) of nAChRs have also been described:the acetylcholine binding protein(AChBP) co-crystallized with agonists and antagonists, and the ECD of α1-nAChRs.<ref>PMID:18987633</ref> | + | X-ray structure of homologues of the extracellular domain(ECD) of nAChRs have also been described:the acetylcholine binding protein(AChBP) co-crystallized with agonists and antagonists, and the ECD of α1-nAChRs. Most pLGICs undergo desensitization on prolonged exposure to agonist, complicating structural investigations of the transient open conformation. <ref>PMID:18987633</ref> |
== Superimpose HAP on AChBP == | == Superimpose HAP on AChBP == | ||
| Line 14: | Line 14: | ||
The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail [http://en.wikipedia.org/wiki/Lymnaea_stagnalis Lymnaea stagnalis]. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref>PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref>PMID:11683996</ref> | The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail [http://en.wikipedia.org/wiki/Lymnaea_stagnalis Lymnaea stagnalis]. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref>PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref>PMID:11683996</ref> | ||
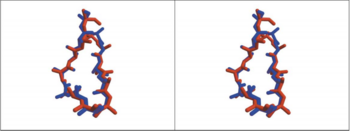
| - | [[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. | + | [[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. 3. Comparison, in Stere, of the 3D Structure of HAP(Red) and Loop 182-193 of AChBP(Blue)]] |
In order to use the complex between α-BTX and HAP to identify the binding site of the AChR, the structure of HAP should homologous with the α-subunits of nAChR. AChBP is a very important and ideal model to study the structure of AChR, which structure has already been solved. So comparing the HAP with the AChBP will show whether HAP can be used as a model to study the binding site of AChR. | In order to use the complex between α-BTX and HAP to identify the binding site of the AChR, the structure of HAP should homologous with the α-subunits of nAChR. AChBP is a very important and ideal model to study the structure of AChR, which structure has already been solved. So comparing the HAP with the AChBP will show whether HAP can be used as a model to study the binding site of AChR. | ||
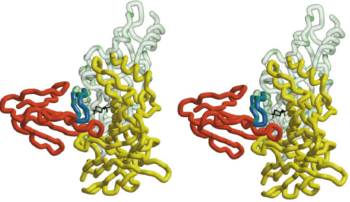
| - | [[Image:Combined BTX HAP and AchBP.png|thumb|350px|Fig. | + | [[Image:Combined BTX HAP and AchBP.png|thumb|350px|Fig. 4. A Stereo View of the Combined Model of α-BTX-HAP(Red) and AChBP subunits]] |
| - | The overly of the first 12 residues of the 13-mer HAP on AChBP residues 182-193 shows that the HAP has almost the same conformation with the loop 182-193 of AChBP(Fig | + | The overly of the first 12 residues of the 13-mer HAP on AChBP residues 182-193 shows that the HAP has almost the same conformation with the loop 182-193 of AChBP(Fig 3), in the figure the red one is 13-mer little peptide and the blue one is loop 182-193 of AChBP. |
| - | The figure | + | The figure 4 shows that the <scene name='68/688431/Btx_complex_with_two_subunits/1'>superposition of the HAP on loop 182-193</scene> the α-BTX to fit exquisitely into the interface of two subunits of the pentameric AChBP. it shows the stereo view of the combined model of α-BTX-HAP(Red) and AChBP structure with subunit A in green and subunit B in yellow showing the insertion of loop 2 of the toxin into the interface of the to subunits. The blue little peptide is HAP, which superimpose on the loop 182-193 of AChBP. In order to identify more clearly that the little 13-mer peptide is actually have almost the same structure with the 182-193 loop with AChBP, we compare two structures: <scene name='68/688431/Btx_complex_with_two_subunits/5'>removing</scene> the HAP form the structure and <scene name='68/688431/Btx_complex_with_two_subunits/6'> |
superposition</scene> the HAP on the AChBP. | superposition</scene> the HAP on the AChBP. | ||
| Line 38: | Line 38: | ||
In nAChR, the ligand-binding site is located at the interface between two subunits. The homopentameric α7 receptor contains five identical ligand binding sites. In these sites acrtylcholine is expected to bind through [http://en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interactions], where the positive charge of the quaternary ammonium of acetylcholine interacts with the electron-rich aromatic side chains.<ref>PMID:11357122</ref> The ACh binding site in AChBP was assigned by the localization of a solvent molecule (positively charge HEPES) seen near residues corresponding to the 187-199 loop of the AChR α subunit and stacking on the corresponding Trp 143.<ref>PMID:11683996</ref> <scene name='68/688431/Hepes_five_subunits/2'>HEPES</scene> can be refined in the current AChBP structure, it does not make any specific hydrogen bonds with the protein, it stacks with its quaternary ammonium onto <scene name='68/688431/Hepes_trp143/1'>Trp 143</scene> making cation-π interactions as expected for nicotinic agonists.<ref>PMID:11357122</ref> | In nAChR, the ligand-binding site is located at the interface between two subunits. The homopentameric α7 receptor contains five identical ligand binding sites. In these sites acrtylcholine is expected to bind through [http://en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interactions], where the positive charge of the quaternary ammonium of acetylcholine interacts with the electron-rich aromatic side chains.<ref>PMID:11357122</ref> The ACh binding site in AChBP was assigned by the localization of a solvent molecule (positively charge HEPES) seen near residues corresponding to the 187-199 loop of the AChR α subunit and stacking on the corresponding Trp 143.<ref>PMID:11683996</ref> <scene name='68/688431/Hepes_five_subunits/2'>HEPES</scene> can be refined in the current AChBP structure, it does not make any specific hydrogen bonds with the protein, it stacks with its quaternary ammonium onto <scene name='68/688431/Hepes_trp143/1'>Trp 143</scene> making cation-π interactions as expected for nicotinic agonists.<ref>PMID:11357122</ref> | ||
| - | The superimposed model of AChBP and α-BTX suggests that the putative agonist HEPES seen in the AChBP structure is blocked from entering or leaving the AChBP interface cleft by the insertion of <scene name='68/688431/Hepes_black_loop_2/1'>loop 2</scene> of α-BTX into that cleft. This clarifies and explains the strong inhibition of AChR function by the toxin.<ref>PMID:11683996</ref> The superposition of the HAP on loop 182-193 of AChBP(Fig. | + | The superimposed model of AChBP and α-BTX suggests that the putative agonist HEPES seen in the AChBP structure is blocked from entering or leaving the AChBP interface cleft by the insertion of <scene name='68/688431/Hepes_black_loop_2/1'>loop 2</scene> of α-BTX into that cleft. This clarifies and explains the strong inhibition of AChR function by the toxin.<ref>PMID:11683996</ref> The superposition of the HAP on loop 182-193 of AChBP(Fig.4)show that the major interaction between α-BTX and AChR α subunit, occur in residues187-192 of that sununit. |
Revision as of 11:38, 2 February 2015
| |||||||||||
Quiz
References
- ↑ Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
- ↑ Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2013 Oct 28. PMID:24167270 doi:http://dx.doi.org/10.1073/pnas.1313156110
- ↑ Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009 Jan 1;457(7225):111-4. Epub 2008 Nov 5. PMID:18987633 doi:10.1038/nature07462
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor
- ↑ Samson AO, Levitt M. Inhibition mechanism of the acetylcholine receptor by alpha-neurotoxins as revealed by normal-mode dynamics. Biochemistry. 2008 Apr 1;47(13):4065-70. doi: 10.1021/bi702272j. Epub 2008 Mar 8. PMID:18327915 doi:http://dx.doi.org/10.1021/bi702272j
Proteopedia Page Contributors and Editors (what is this?)
Ma Zhuang, Zicheng Ye, Angel Herraez, Alexander Berchansky, Michal Harel