This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Z-DNA
From Proteopedia
| Line 28: | Line 28: | ||
Z-DNA binding proteins have common structural characteristics. The binding domains of these proteins can substitute one another and thus can act as competitive inhibitors against one another. As explained above, disruption in the Z-DNA binding region of E3L reduces its pathogenicity. All these observations are important pointers towards the biological importance of Z-DNA.<ref name ='Wang'>PMID:17485386</ref> | Z-DNA binding proteins have common structural characteristics. The binding domains of these proteins can substitute one another and thus can act as competitive inhibitors against one another. As explained above, disruption in the Z-DNA binding region of E3L reduces its pathogenicity. All these observations are important pointers towards the biological importance of Z-DNA.<ref name ='Wang'>PMID:17485386</ref> | ||
| + | |||
| + | === High-resolution crystal structure of Z-DNA in complex with Cr3+ cations <ref>DOI 10.1007/s00775-015-1247-5</ref>=== | ||
| + | |||
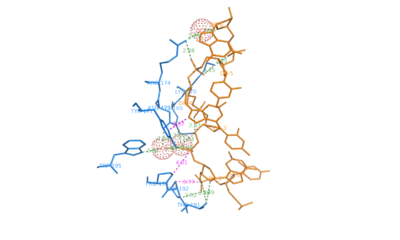
| + | Trivalent chromium, a d<sup>3</sup> cation, is poorly taken up by living cells. The Cr<sup>3+</sup> ions are the final product of in vivo Cr<sup>6+</sup> metabolism. However, Cr<sup>3+</sup> in contrast to Cr<sup>6+</sup> can form coordination complexes with macromolecules in the cells. In vitro biochemical experiments have shown that exposure of cells to Cr6+ yields binary (DNA–Cr<sup>3+</sup>) and ternary (DNA–Cr<sup>3+</sup>–ligand) adducts, DNA crosslinks, as well as oxidative DNA lesions. Despite the interest in DNA–Cr<sup>3+</sup> interactions in biological systems, the existing literature provides detailed crystallographic structural data for only two, low-resolution DNA–Cr<sup>3+</sup>:DNA polymerase-β complexes, PDB [[1zqe]] (3.7 Å) ֵand [[1huz]] (2.6 ֵÅ). | ||
| + | |||
| + | Our work is part of our project aimed at characterizing metal-binding properties of left-handed Z-DNA helices. The three Cr3+ cations found in the asymmetric unit of the d(CGCGCG)<sub>2</sub>–Cr<sup>3+</sup> crystal structure do not form direct coordination bonds with either the guanine N/O atoms or the phosphate groups of the Z-DNA. <scene name='69/693575/Cv/6'>Note the alternate conformations</scene> (<span style="color:lime;background-color:black;font-weight:bold;">I, green</span>; <span style="color:orange;background-color:black;font-weight:bold;">II, orange</span>) along the DNA chains. <scene name='69/693575/Cv/7'>Click here to see the animation of this scene</scene>. <font color='darkmagenta'><b>Cr<sup>3+</sup> cations shown as purple spheres</b></font>. Instead, only water-mediated contacts between the nucleic acid and the Cr<sup>3+</sup> cations are observed. The coordination spheres of Cr<sup>3+</sup>(1) and Cr<sup>3+</sup>(2) contain six water molecules each. The Cr<sup>3+</sup>(1) and Cr<sup>3+</sup>(2) ions are bridged by three water molecules from their coordination spheres, one of which (Wat1) is split into two sites. The hydration patterns of Cr<sup>3+</sup>(1) and Cr<sup>3+</sup>(2) are <scene name='69/693575/Cv/8'>irregular and difficult to define</scene> (<font color='red'><b>water molecules are represented by red spheres</b></font>). The Cr<sup>3+</sup>(3) cation has <scene name='69/693575/Cv/9'>distorted square pyramidal geometry</scene>. | ||
| + | |||
| + | We have used Z-DNA crystals to obtain accurate information about the geometrical parameters characterizing the coordination of Cr3+ ions by left-handed Z-DNA. The d(CGCGCG)2–Cr<sup>3+</sup> structure is an excellent illustration of the flexibility of the Z-DNA molecule, visible in the adoption of multiple conformations (by the phosphate groups and the G2 nucleotide), in response to changes in its electrostatic and hydration environment, caused by the introduction of hydrated metal complexes. | ||
</StructureSection> | </StructureSection> | ||
__NOTOC__ | __NOTOC__ | ||
Revision as of 12:58, 25 February 2015
| |||||||||||
Movie Depicting ADAR1 binding to Z-DNA
Comparison of the three helices and helical parameters of DNA
Sources[7]
|
|
|
| Parameter | A-DNA | B-DNA | Z-DNA |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Residues per turn | 11 | 10.5 | 12 |
| Axial rise [Å] | 2.55 | 3.4 | 3.7 |
| Helix pitch(°) | 28 | 34 | 45 |
| Base pair tilt(°) | 20 | −6 | 7 |
| Rotation per residue (°) | 33 | 36 | -30 |
| Diameter of helix [Å] | 23 | 20 | 18 |
| Glycosidic bond configuration dA,dT,dC dG | anti anti | anti anti | anti syn |
| Sugar pucker dA,dT,dC dG | C3'-endo C3'-endo | C2'-endo C2'-endo | C2'-endo C3'-endo |
| Intrastrand phosphate-phosphate distance [Å] dA,dT,dC dG | 5.9 5.9 | 7.0 7.0 | 7.0 5.9 |
| Sources:[8][9][10] | |||
3D structures of Z-DNA
Updated on 25-February-2015
1woe, 3qba – ZDNA hexamer CGCGCG
1qbj – ZDNA hexamer CGCGCG + hADAR1 Zα domain - human
2heo - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zα domain – mouse
2eyi - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zβ domain
1j75 - ZDNA hexamer CGCGCG + DLM-1 Zα domain
Additional Resources
For additional information, see: Nucleic Acids
References
- ↑ 1.0 1.1 1.2 Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003 Jul;4(7):566-72. PMID:12838348 doi:10.1038/nrg1115
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007 May 1;12:4424-38. PMID:17485386
- ↑ Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005 Oct 20;437(7062):1183-6. PMID:16237447 doi:10.1038/nature04088
- ↑ Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999 Jun 11;284(5421):1841-5. PMID:10364558
- ↑ Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1514-8. Epub 2004 Feb 2. PMID:14757814 doi:10.1073/pnas.0308260100
- ↑ Drozdzal P, Gilski M, Kierzek R, Lomozik L, Jaskolski M. High-resolution crystal structure of Z-DNA in complex with Cr cations. J Biol Inorg Chem. 2015 Feb 17. PMID:25687556 doi:http://dx.doi.org/10.1007/s00775-015-1247-5
- ↑ http://203.129.231.23/indira/nacc/
- ↑ Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791-846. PMID:6383204 doi:http://dx.doi.org/10.1146/annurev.bi.53.070184.004043
- ↑ Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680-6. PMID:514347
- ↑ Sinden, Richard R (1994-01-15). DNA structure and function (1st ed.). Academic Press. pp. 398. ISBN 0-12-645750-6.
Proteopedia Page Contributors and Editors (what is this?)
Adithya Sagar, Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Donald Voet, Jaime Prilusky, Karl Oberholser, Eran Hodis