We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1066
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
= Mechanism = | = Mechanism = | ||
| + | |||
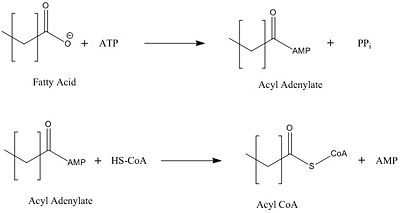
| + | == General mechanism for the activation of fatty acids == | ||
| + | |||
| + | [[Image:acyl coa synthetase.jpg|400 px|center|thumb|Figure Legend]] | ||
== Structural basis for housing lipid substrates longer than the enzyme == | == Structural basis for housing lipid substrates longer than the enzyme == | ||
| Line 16: | Line 20: | ||
The ability for FadD13 to transport and activate fatty acids of the maximum tested length C26, lies in it being a peripheral membrane protein. FadD13 is attached to the membrane via electrostatic interactions in the N-terminal domain. Of importance in the region is the arginine rich lid-loop which serves to block the transport of fatty acids into the enzyme. Once the lid-loop is opened, fatty acids may be pulled from the membrane into a hydrophobic tunnel, which is the main structural component by which fatty acids are transported from the membrane into the cell. Negatively charged residues at the active site of FadD13 are the driving factor in the attraction of the fatty acid from the membrane through the hydrophobic tunnel of the enzyme. | The ability for FadD13 to transport and activate fatty acids of the maximum tested length C26, lies in it being a peripheral membrane protein. FadD13 is attached to the membrane via electrostatic interactions in the N-terminal domain. Of importance in the region is the arginine rich lid-loop which serves to block the transport of fatty acids into the enzyme. Once the lid-loop is opened, fatty acids may be pulled from the membrane into a hydrophobic tunnel, which is the main structural component by which fatty acids are transported from the membrane into the cell. Negatively charged residues at the active site of FadD13 are the driving factor in the attraction of the fatty acid from the membrane through the hydrophobic tunnel of the enzyme. | ||
| - | + | ||
= Structure = | = Structure = | ||
== General overview == | == General overview == | ||
Revision as of 01:39, 1 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644