Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== '''Structure''' == | == '''Structure''' == | ||
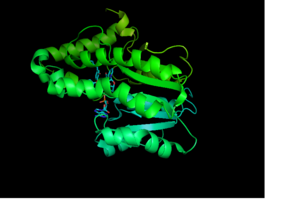
| - | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> with separate ligand binding sites in each subunit. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit features a single domain with a central core that supports a NADH binding site. The alpha6 and alpha7 helices of the monomer form one side of the fatty acyl substrate binding crevice of InhA. Within this binding crevice, many hydrophobic side chains from the amino acids involved in forming the substrate binding loop, <scene name='69/694241/Monomer_subunit_196_219/1'> residues 196-219</scene>, interact with the substrates. One side of this cavity is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to distinguish between shorter and longer substrates and provide specificity for the enoyl-ACP reductase reaction. | + | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> with separate ligand binding sites in each subunit. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit features a single domain with a central core that supports a NADH binding site. The alpha6 and alpha7 helices of the monomer form one side of the fatty acyl substrate binding crevice of InhA. Within this binding crevice, many hydrophobic side chains from the amino acids involved in forming the substrate binding loop, <scene name='69/694241/Monomer_subunit_196_219/1'> residues 196-219</scene>, interact with the substrates. One side of this cavity is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to distinguish between shorter and longer substrates and provide <scene name='69/694241/Substrate_binding_pocket/1'>specificity</scene> for the enoyl-ACP reductase reaction. |
| - | <scene name='69/694241/Substrate_binding_pocket/1'>substrate binding pocket</scene> | ||
[[Image:Monomer_Publication_Image.png |300 px|left|thumb|<div align="center">InhA Monomer with NAD and 2TK]] | [[Image:Monomer_Publication_Image.png |300 px|left|thumb|<div align="center">InhA Monomer with NAD and 2TK]] | ||
<scene name='69/694241/Monomer_subunit/2'>NADH binding site</scene>. | <scene name='69/694241/Monomer_subunit/2'>NADH binding site</scene>. | ||
| - | |||
| - | |||
| - | <scene name='69/694241/Sbl_amino_acids/1'> residues 196-219</scene> | ||
| - | |||
| - | <scene name='69/694241/Sbl_inside_view/1'>substrate binding loop</scene> | ||
| - | |||
| - | |||
Revision as of 02:41, 7 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||