We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1075
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
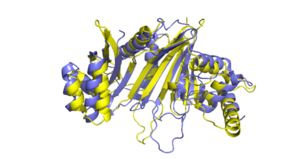
[[Image:EspG5_and_EspG3_overlap_W.png|300 px|right|thumb|EspG5 and EspG3]] | [[Image:EspG5_and_EspG3_overlap_W.png|300 px|right|thumb|EspG5 and EspG3]] | ||
| + | |||
| + | <scene name='69/694242/Specific_contact_residues/1'>TextToBeDisplayed</scene> | ||
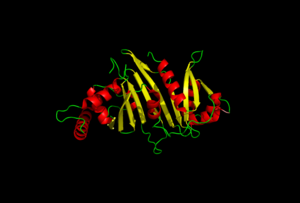
| + | The random loop on the cigar shaped PE-PPE ligand binds to the B2-B3 sheets on this EspG protein. | ||
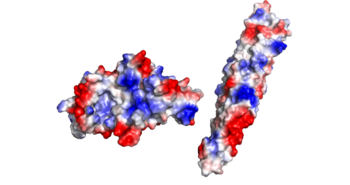
The residues on the random turn in the PE-PPE protein are key for the specificity to the EspG protein. Coils between PE-PPE proteins vary greatly and influence binding affinity. Combined with its β-2 & β-3 interactions on the EspG protein, the EspG protein is PE-PPE specific. These make up the bulk of residue interactions in the complex. | The residues on the random turn in the PE-PPE protein are key for the specificity to the EspG protein. Coils between PE-PPE proteins vary greatly and influence binding affinity. Combined with its β-2 & β-3 interactions on the EspG protein, the EspG protein is PE-PPE specific. These make up the bulk of residue interactions in the complex. | ||
Revision as of 12:43, 7 April 2015
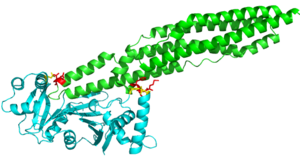
Binding Specificity of EspG Proteins to PE-PPE Proteins in mycobacterium tuberculosis
| |||||||||||
References
- ↑ Ekiert DC, Cox JS. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14758-63. doi:, 10.1073/pnas.1409345111. Epub 2014 Oct 1. PMID:25275011 doi:http://dx.doi.org/10.1073/pnas.1409345111