Sandbox Reserved 1058
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
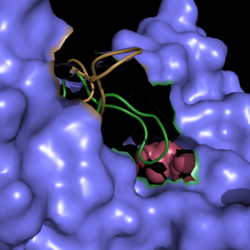
[[Image:Active Loop Shift.png|250 px|center|thumb|'''Figure 1. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. ]] | [[Image:Active Loop Shift.png|250 px|center|thumb|'''Figure 1. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. ]] | ||
The active site loop unbound is represented in green while the active site loop bound is shown in tan in Figure 1. | The active site loop unbound is represented in green while the active site loop bound is shown in tan in Figure 1. | ||
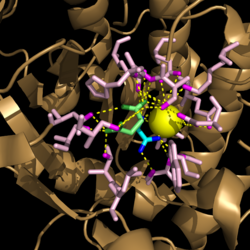
| + | [[Image:Active_Site_Hydrogen_Bonding.png|250 px|left|thumb|'''Figure 3. Active site residues hydrogen bound to a cofactor and the products of the catalyzed isocitrate reaction.''' Glyoxylate is shown in blue, succinate is shown in green, and the Mg<sup>2+</sup> cofactor is shown in yellow.]] | ||
===Inhibitors=== | ===Inhibitors=== | ||
| Line 20: | Line 21: | ||
==Mechanism of Action== | ==Mechanism of Action== | ||
[[Image:TCA_Cycle.png|500 px|left|thumb|'''Figure 2. Citric Acid Cycle with Glyoxylate Shunt Pathway.''' In several bacterial species, there is a carbon conserving gloxylate shunt pathway that converts isocitrate to malate in two steps instead of the usual five steps.]] | [[Image:TCA_Cycle.png|500 px|left|thumb|'''Figure 2. Citric Acid Cycle with Glyoxylate Shunt Pathway.''' In several bacterial species, there is a carbon conserving gloxylate shunt pathway that converts isocitrate to malate in two steps instead of the usual five steps.]] | ||
| - | + | ||
==Disease Association== | ==Disease Association== | ||
===Clinical Implications=== | ===Clinical Implications=== | ||
Revision as of 19:32, 7 April 2015
Isocitrate Lyase from Mycobacterium tuberculosis
| |||||||||||