Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== '''General Structural Information''' == | == '''General Structural Information''' == | ||
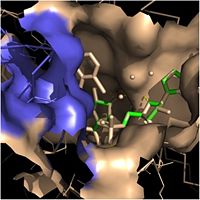
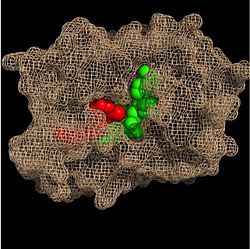
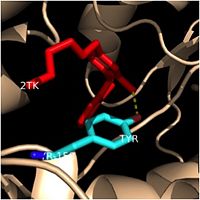
| - | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit is composed of 289 residues and features a typical [http://en.wikipedia.org/wiki/Rossmann_fold] containing a single NADH binding site. The <scene name='69/694241/Secondary_structure_black/1'>secondary structure</scene> of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white). This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate (2TK) needed to synthesize mycolic acid precursors. The <scene name='69/694241/Helix6_helix7_updated/1'>alpha-6 and alpha-7 helices</scene> of the InhA form one side of the fatty acyl binding crevice, referred to as the <scene name='69/694241/Monomer_subunit_196_219/1'> substrate binding loop</scene> (residues 196-219). [[Image:Fatty Acyl Binding Crevice.jpg|thumb|200px|left|Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates inside)]] | + | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit is composed of 289 residues and features a typical [http://en.wikipedia.org/wiki/Rossmann_fold] containing a single NADH binding site. The <scene name='69/694241/Secondary_structure_black/1'>secondary structure</scene> of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white). This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate (2TK) needed to synthesize mycolic acid precursors. The <scene name='69/694241/Helix6_helix7_updated/1'>alpha-6 and alpha-7 helices</scene> of the InhA form one side of the fatty acyl binding crevice, referred to as the <scene name='69/694241/Monomer_subunit_196_219/1'> substrate binding loop</scene> (residues 196-219). [[Image:Fatty Acyl Binding Crevice.jpg|thumb|200px|left|Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates pictured inside the crevice)]] |
Talk generally about catalytic triad. More specific details in sections below. | Talk generally about catalytic triad. More specific details in sections below. | ||
Revision as of 20:00, 7 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||