Introduction
FAS-II System
Mechanism of Action
General Structural Information
Crystal structures of InhA reveal a (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit. Each subunit is composed of 289 residues and features a typical
Rossmann fold containing a single NADH binding site. The of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white). This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate needed to synthesize mycolic acid precursors. The of the InhA form one side of the fatty acyl binding crevice, referred to as the (residues 196-219).
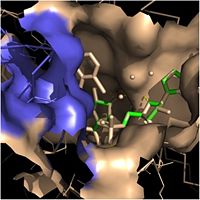
Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates pictured inside the crevice)
One side of this crevice is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to select for fatty acyl chains longer than 16 carbons to successfully produce mycolic acid precursors.
Ligands
The two ligands that play a role in the function of InhA are NADH and the long-chain fatty acyl substrate. NADH, or nicotinamide adenine dinucleotide, is a cofactor found in all living cells. It is especially important in the function of InhA because it helps position the fatty acyl substrate within the fatty acyl binding crevice, thereby improving the overall activity of InhA. Once properly positioned, the fatty acyl substrates are reduced at the trans double bond between C2 and C3, forming mycolic acid precursors that eventually compose the cell wall of mycobacteria.
Fatty Acyl Binding Crevice
Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Due to its position within the crevice, the
trans double bond of the fatty acyl substrate is found near the closed end of the crevice and is located directly adjacent to the nicotinamide ring of
NADH.
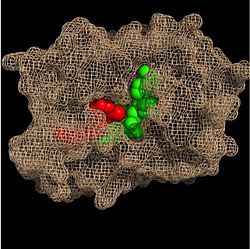
Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)
These two molecules interact with each other via hydrogen bonding. The first hydrogen bond occurs between the phosphate oxygen of NADH and the amide nitrogen of the N-acetylcysteamine portion of the fatty acyl substrate. The second hydrogen bond occurs between the 2'-hydroxyl of the nicotinamide ribose of NADH and the thioester carbonyl oxygen of the fatty acyl substrate.
Substrate Binding Loop Flexibility
In addition to the hydrogen bonding that occurs between the NADH and fatty acyl substrate within the crevice, each of these molecules is also held in place within the crevice through interactions with the side chains of surrounding (purple) residues. The majority of these residues anchoring the substrates are found within the substrate binding loop itself, including Ala-198, Met-199, Ala-201, Ile-202, Leu-207, Ile-215, and Leu-218. Additional hydrophobic amino acids that are not a part of the substrate binding loop also play a role in positioning and stabilizing the fatty acyl chain in the crevice. Studies have found that the fatty acyl substrate adopts a u-shaped conformation to facilitate binding. As the fatty acyl substrate binds in the crevice, the substrate binding loop shifts outward toward the solvent, and the Tyr-158 residue is rotated to facilitate the binding of fatty acyl chains of 16 carbons or greater. During this conformational change upon substrate binding, no hydrogen bonds are broken, which supports the flexibility of the substrate binding loop. It is likely that this flexibility of the substrate binding loop provides increased freedom for the active site to accept fatty acyl substrates of varying carbon chain lengths.
Importance of Tyr-158
One example of a hydrophobic amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is
Tyr-158.
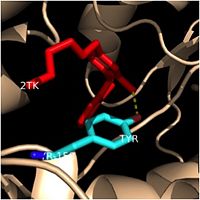
Tyr-158 (turquoise) hydrogen bonding to the fatty acyl substrate, 2TK (red)
This amino acid is conserved in other enoyl-ACP reductases in both bacteria and plants, so it likely plays an essential role in the function of these specific enzymes. Studies have shown that Tyr-158 forms the only direct hydrogen bond that exists between the InhA protein and the fatty acyl substrate. This hydrogen bond occurs between the hydroxyl oxygen on the side chain of Tyr-158 and the thioester carbonyl oxygen of the fatty acyl substrate. As a result of the highly conserved nature and the specific hydrogen bonding capabilities of this amino acid, it is likely that Tyr-158 plays an important role in fatty acyl substrate binding in enoyl-ACP reductases.
Catalytic Triad
Structural studies have shown that InhA possesses the , composed of Phe-149, Tyr-158, and Lys-165. This catalytic triad of InhA is analogous to the classic Ser-Tyr-Lys catalytic triad of the SDR (Short-chain Dehydrogenase Reductase) family in terms of position and function within the enzyme. As discussed previously, the likely role of Tyr-158 is to position the fatty acyl substrate within the fatty acyl binding crevice. Second, the side chain of the Lys-165 residue in InhA functions similarly to the catalytic Lys residues in other SDR enzymes by interacting with the 3'-hydroxyl of the nicotinamide ring of NADH to hold this cofactor in place within the fatty acyl binding crevice. Finally, the exact role of the catalytic Phe-149 residue in InhA is unknown. It has been hypothesized to play a role in helping the fatty acyl substrate adopt its desired u-shape prior to binding. However, it is more widely believed that this catalytic residue merely distinguishes InhA as a dehydrogenase.
Clinical Applications



