We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1073
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
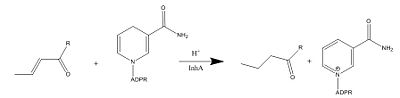
The InhA gene encodes for the InhA protein. InhA catalyzes the NADH-dependent reduction of the trans double bond between positions C2-C3 of fatty acyl substrates. InhA prefers fatty acyl substrates of C16 or longer, which is consistent of the protein being a member of the FAS-II system. The longer chain length specificity of InhA distinguishes the enzyme from other enoyl-ACP reductase analogues. | The InhA gene encodes for the InhA protein. InhA catalyzes the NADH-dependent reduction of the trans double bond between positions C2-C3 of fatty acyl substrates. InhA prefers fatty acyl substrates of C16 or longer, which is consistent of the protein being a member of the FAS-II system. The longer chain length specificity of InhA distinguishes the enzyme from other enoyl-ACP reductase analogues. | ||
| - | + | [[Image:Mechanism-InhA.JPG|thumb|400px|Figure 1. Mechanism of InhA protein]] | |
== Structure == | == Structure == | ||
| Line 31: | Line 31: | ||
Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser<sup>94</sup> to Ala of InhA was sufficient enough to have isoniazid resistance. | Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser<sup>94</sup> to Ala of InhA was sufficient enough to have isoniazid resistance. | ||
| - | + | === Other Inhibitors === | |
| + | |||
| + | Drug resistance of ''M. tuberculosis''has become a huge problem for the development of antibiotics. A drug screen of potential inhibitors of InhA (300 compounds), were composed of inhibitors of the ''P. falciparum'' | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 21:14, 8 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA
| |||||||||||