Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== '''General Structural Information''' == | == '''General Structural Information''' == | ||
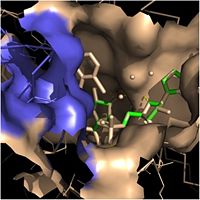
| - | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit <ref name="InhA"> Rozwarski, D.A. (1999). Crystal Structure of the ''Mycobacterium tuberculosis'' Enoyl-ACP Reductase, InhA, in Complex with NAD<sup>+</sup> and a C16 Fatty Acyl Substrate. ''Journal of Biological Chemistry, 274(22),'' 15582-15589. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/10336454 10336454] DOI: [http://www.ncbi.nlm.nih.gov/pubmed/10336454 10.1074/jbc.274.22.15582]</ref>. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit is composed of 289 residues and features a typical [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann fold] containing a single NADH binding site. The <scene name='69/694241/Secondary_structure_black/1'>secondary structure</scene> of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white) <ref name="Phe149"> Bell, A.F. (2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. ''Journal of the American Chemical Society, 129,'' 6425-6431. DOI: [http://pubs.acs.org/doi/abs/10.1021/ja068219m 10.1021/ja068219m]</ref>. This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate needed to synthesize mycolic acid precursors. [[Image:Fatty Acyl Binding Crevice.jpg|thumb|200px|left|Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates pictured inside the crevice)]] The <scene name='69/694241/Helix6_helix7_updated/1'>alpha-6 and alpha-7 helices</scene> of the InhA form one side of the fatty acyl binding crevice, referred to as the <scene name='69/694241/Monomer_subunit_196_219/1'> substrate binding loop</scene> (residues 196-219). One side of this crevice is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to select for fatty acyl chains longer than 16 carbons to successfully produce mycolic acid precursors <ref name="InhA" />. | + | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene> (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit <ref name="InhA"> Rozwarski, D.A. (1999). Crystal Structure of the ''Mycobacterium tuberculosis'' Enoyl-ACP Reductase, InhA, in Complex with NAD<sup>+</sup> and a C16 Fatty Acyl Substrate. ''Journal of Biological Chemistry, 274(22),'' 15582-15589. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/10336454 10336454] DOI: [http://www.ncbi.nlm.nih.gov/pubmed/10336454 10.1074/jbc.274.22.15582]</ref>. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit is composed of 289 residues and features a typical [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann fold] containing a single NADH binding site. The <scene name='69/694241/Secondary_structure_black/1'>secondary structure</scene> of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white) <ref name="Phe149"> Bell, A.F. (2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. ''Journal of the American Chemical Society, 129,'' 6425-6431. DOI: [http://pubs.acs.org/doi/abs/10.1021/ja068219m 10.1021/ja068219m]</ref>. This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate needed to synthesize mycolic acid precursors. [[Image:Fatty Acyl Binding Crevice.jpg|thumb|200px|left|Figure 2. Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates pictured inside the crevice)]] The <scene name='69/694241/Helix6_helix7_updated/1'>alpha-6 and alpha-7 helices</scene> of the InhA form one side of the fatty acyl binding crevice, referred to as the <scene name='69/694241/Monomer_subunit_196_219/1'> substrate binding loop</scene> (residues 196-219). One side of this crevice is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to select for fatty acyl chains longer than 16 carbons to successfully produce mycolic acid precursors <ref name="InhA" />. |
==='''Ligands'''=== | ==='''Ligands'''=== | ||
| Line 23: | Line 23: | ||
== '''Fatty Acyl Binding Crevice''' == | == '''Fatty Acyl Binding Crevice''' == | ||
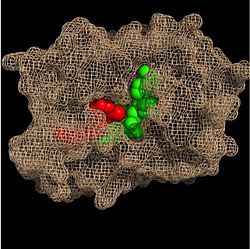
| - | Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Due to its position within the crevice, the ''trans'' double bond of the fatty acyl substrate is found near the closed end of the crevice and is located directly adjacent to the nicotinamide ring of [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NADH]. [[Image:Binding Pocket - Mesh.jpg|thumb|250px|right|Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)]] These two molecules interact with each other via hydrogen bonding. The first hydrogen bond occurs between the phosphate oxygen of NADH and the amide nitrogen of the N-acetylcysteamine portion of the fatty acyl substrate. The second hydrogen bond occurs between the 2'-hydroxyl of the nicotinamide ribose of NADH and the thioester carbonyl oxygen of the fatty acyl substrate <ref name="InhA" />. | + | Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Due to its position within the crevice, the ''trans'' double bond of the fatty acyl substrate is found near the closed end of the crevice and is located directly adjacent to the nicotinamide ring of [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NADH]. [[Image:Binding Pocket - Mesh.jpg|thumb|250px|right|Figure 3. Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)]] These two molecules interact with each other via hydrogen bonding. The first hydrogen bond occurs between the phosphate oxygen of NADH and the amide nitrogen of the N-acetylcysteamine portion of the fatty acyl substrate. The second hydrogen bond occurs between the 2'-hydroxyl of the nicotinamide ribose of NADH and the thioester carbonyl oxygen of the fatty acyl substrate <ref name="InhA" />. |
==='''Substrate Binding Loop Flexibility''' === | ==='''Substrate Binding Loop Flexibility''' === | ||
| Line 31: | Line 31: | ||
==='''Importance of Tyr-158'''=== | ==='''Importance of Tyr-158'''=== | ||
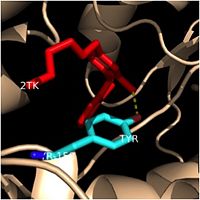
| - | One example of a hydrophobic amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is [[Tyr-158]]. [[Image:Tyr-158.jpg|thumb|200px|left|Tyr-158 (light blue) hydrogen bonding to the fatty acyl substrate, 2TK (red)]] This amino acid is conserved in other enoyl-ACP reductases in both bacteria and plants, so it likely plays an essential role in the function of these specific enzymes. Studies have shown that Tyr-158 forms the only direct hydrogen bond that exists between the InhA protein and the fatty acyl substrate. This hydrogen bond occurs between the hydroxyl oxygen on the side chain of Tyr-158 and the thioester carbonyl oxygen of the fatty acyl substrate. Consequently, the Tyr-158 residue acts to stabilize the enolate intermediate that forms during the hydride transfer reaction <ref name="InhA" />. | + | One example of a hydrophobic amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is [[Tyr-158]]. [[Image:Tyr-158.jpg|thumb|200px|left|Figure 4. Tyr-158 (light blue) hydrogen bonding to the fatty acyl substrate, 2TK (red)]] This amino acid is conserved in other enoyl-ACP reductases in both bacteria and plants, so it likely plays an essential role in the function of these specific enzymes. Studies have shown that Tyr-158 forms the only direct hydrogen bond that exists between the InhA protein and the fatty acyl substrate. This hydrogen bond occurs between the hydroxyl oxygen on the side chain of Tyr-158 and the thioester carbonyl oxygen of the fatty acyl substrate. Consequently, the Tyr-158 residue acts to stabilize the enolate intermediate that forms during the hydride transfer reaction <ref name="InhA" />. |
Revision as of 22:29, 8 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Rozwarski, D.A. (1999). Crystal Structure of the Mycobacterium tuberculosis Enoyl-ACP Reductase, InhA, in Complex with NAD+ and a C16 Fatty Acyl Substrate. Journal of Biological Chemistry, 274(22), 15582-15589. PMID: 10336454 DOI: 10.1074/jbc.274.22.15582
- ↑ 2.0 2.1 2.2 Bell, A.F. (2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. Journal of the American Chemical Society, 129, 6425-6431. DOI: 10.1021/ja068219m