Dioxygenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection | + | <StructureSection size="450" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> |
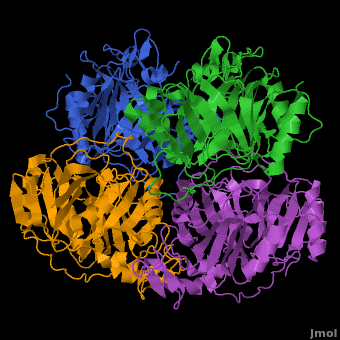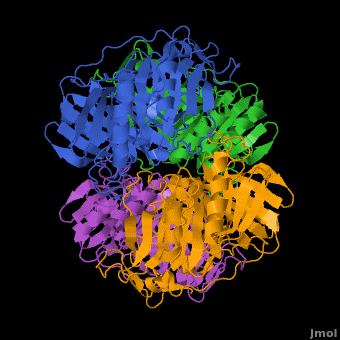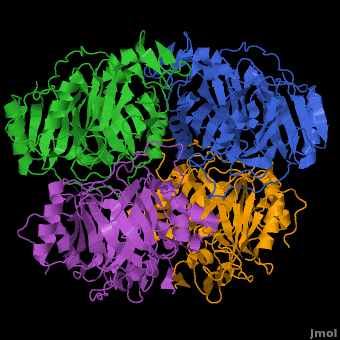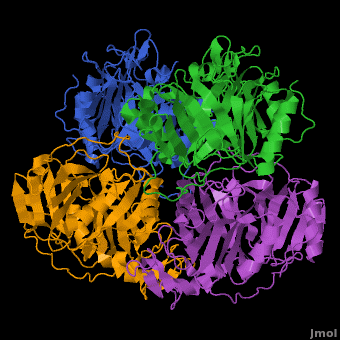
'''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> | '''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> | ||
Revision as of 10:54, 7 May 2015
| |||||||||||
3D structures of Protocatechuate 3,4-dioxygenase
Updated on 07-May-2015
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky