Sandbox Reserved 1059
From Proteopedia
| Line 1: | Line 1: | ||
=='''NrdH of ''Mycobacterium tuberculosis''''' == | =='''NrdH of ''Mycobacterium tuberculosis''''' == | ||
| - | ===Introduction=== | + | ==='''Introduction'''=== |
| - | NrdH | + | NrdH redoxins are small reductases, and they contain similar amino acid sequences to [http://en.wikipedia.org/wiki/Glutaredoxin glutaredoxins] and [http://en.wikipedia.org/wiki/Mycoredoxin mycoredoxins]. However, NrdH redoxins are different because of their [http://en.wikipedia.org/wiki/Thioredoxin thioredoxin]-like activity. Their main function is to act as the electron donor for class 1b [http://en.wikipedia.org/wiki/Ribonucleotide_reductase ribonucleotide reductases], which are important for the conversion of [http://en.wikipedia.org/wiki/Ribonucleotide ribonucleotides] to [http://en.wikipedia.org/wiki/Deoxyribonucleotide deoxyribonucleotides] <ref name="Laer">DOI: 10.1074/jbc.M112.392688</ref>. The process of ribonucleotide reduction is one of the most fundamental biochemical processes that is required for the existence of DNA-based life. It is the only de novo pathway to synthesize deoxyribonucleotides. Deoxyribonucleotides are the building blocks of DNA and are synthesized from ribonucleotides by reducing the 2’OH in a radical based reaction. The deoxyribonucleotides are then used as precursors for the process of DNA synthesis. This reaction is catalyzed by ribonucleotide reductases <ref name="Phulera">DOI: 10.1021/bi400191z</ref>. |
| - | + | ==='''Ribonucleotide Reductases (RNRs)'''=== | |
| + | All RNRs are related because they are necessary for all living cells, with the exception of a couple types of parasites and obligate endosymbionts. This is evident due to the catalytic core, which is structurally conserved across all extant RNRs. RNRs are essential for the processes of DNA replication and repair <ref name="Lundin">DOI: 10.1186/1471-2148-10-383</ref>. | ||
| - | There are three classes of | + | There are three classes of RNRs. These classes are divided based on the mechanism of radical generation in the reaction. |
[[Image:Classes_of_ribonucleotide_reductases_(RNR).png]] | [[Image:Classes_of_ribonucleotide_reductases_(RNR).png]] | ||
| + | Class I enzymes reduce nucleotide 5’-diphosphates, while the other 2 classes reduce ribonucleotide 5’-triphosphates <ref name="Phulera" />. Class I RNRs are oxygen-dependent enzymes that contain a di-iron cluster <ref name="Laer" />. | ||
| - | Class | + | Class 1 RNRs generate a [http://en.wikipedia.org/wiki/Tyrosine tyrosyl] [http://en.wikipedia.org/wiki/Radical_(chemistry) radical] in another subunit, which is NrdB for class 1a and NrdF in class 1b. The tyrosyl radical is then transferred to the catalytic subunit, which is NrdA in class Ia and NrdE in class 1b <ref name="Lundin" />. |
| - | + | ||
| - | + | ||
At the end of each cycle of ribonucleotide reduction, the ribonucleotide reductase needs to be reduced in order to be ready for the next reduction cycle. For a class 1a RNR, an external cofactor, such as a glutaredoxin or thioredoxin, performs this reduction step. For class 1b RNRs, this cofactor is known as NrdH. NrdH contains a glutaredoxin-like sequence but behaves like a thioredoxin <ref name="Phulera" />. | At the end of each cycle of ribonucleotide reduction, the ribonucleotide reductase needs to be reduced in order to be ready for the next reduction cycle. For a class 1a RNR, an external cofactor, such as a glutaredoxin or thioredoxin, performs this reduction step. For class 1b RNRs, this cofactor is known as NrdH. NrdH contains a glutaredoxin-like sequence but behaves like a thioredoxin <ref name="Phulera" />. | ||
| + | ==='''NrdH of ''Mycobacterium tuberculosis'''''=== | ||
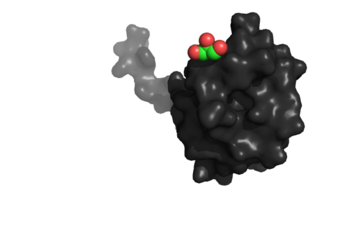
<Structure load='4K8M' size='350' frame='true' align='right' caption='NrdH of ''Mycobacterium tuberculosis''' scene='Insert optional scene name here' /> | <Structure load='4K8M' size='350' frame='true' align='right' caption='NrdH of ''Mycobacterium tuberculosis''' scene='Insert optional scene name here' /> | ||
| + | |||
| + | NrdH is a redox protein, and it is part of a family of redox proteins. The other proteins that maintain the redox balance of NrdH are three thioredoxin and three glutaredoxin-like proteins. Prokaryotes typically maintain redox homeostasis through low-molecular weight thiols (glutathione) and through proteins involved in disulfide exchange (thioredoxins). | ||
[[Image:Image_2_(2).png|350px|left|thumb|Aromatic Amino Acids binding site]] | [[Image:Image_2_(2).png|350px|left|thumb|Aromatic Amino Acids binding site]] | ||
| - | + | NrdH is found in many types of bacteria, such as [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis ''Mycobacterium tuberculosis'']. This bacteria causes the disease [http://en.wikipedia.org/wiki/Tuberculosis tuberculosis], which is a fatal disease if not treated properly. Tuberculosis was the leading cause of death in the United States in the past, and it can be spread through the air from one person to another by coughing, sneezing, or speaking <ref>DOI: 10.1016/j.tube.2003.08.003</ref>. | |
| - | + | ||
== Structure == | == Structure == | ||
| Line 31: | Line 33: | ||
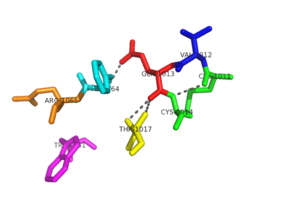
[[Image:Conserved Motifs.png|300px|left|thumb|Conserved motifs]] | [[Image:Conserved Motifs.png|300px|left|thumb|Conserved motifs]] | ||
| - | Within the <scene name='69/694226/Cvqc_motif/1'>CVQC motif</scene>, the amide oxygen of glutamine residue is firmly hydrogen bonded with the peptidyl nitrogen of Phe-44. The amide nitrogen of glutamine is then available for further hydrogen bonding. The carbonyl oxygen of Val-12 hydrogen bonds with peptidyl nitrogen of Ala-16. The residues between the two cysteines are known to affect redox potentials and pKa values. Also, by changing the target proteins, in turn, they regulate the function | + | Within the <scene name='69/694226/Cvqc_motif/1'>CVQC motif</scene>, the amide oxygen of glutamine residue is firmly hydrogen bonded with the peptidyl nitrogen of Phe-44. The amide nitrogen of glutamine is then available for further hydrogen bonding. The carbonyl oxygen of Val-12 hydrogen bonds with peptidyl nitrogen of Ala-16. This hydrogen bonding leads to stability within the redox active site of NrdH. The N-terminal cysteine of the CVQC motif acts as a nucleophile, whereas the C-terminal cysteine acts as the resolving cysteine. The residues between the two cysteines are known to affect redox potentials and pKa values. Also, by changing the target proteins, in turn, they regulate the function <ref name="Phulera" />. |
The <scene name='69/694226/Wsgfrp_conserved_motif/1'>WSGFRP motif</scene> is stabilized by glutamine of the CVQC motif and phenylalanine is exposed to the solvent. Phe-64 and Val-12 with Ala-16 and Ala-20 create a distinct hydrophobic patch that is exposed to the solvent. This patch is of functional significance that could potentially interact with the C-terminus of RNR. This hydrogen bonding network lends to the stability of the redox active site <ref name="Phulera" />. | The <scene name='69/694226/Wsgfrp_conserved_motif/1'>WSGFRP motif</scene> is stabilized by glutamine of the CVQC motif and phenylalanine is exposed to the solvent. Phe-64 and Val-12 with Ala-16 and Ala-20 create a distinct hydrophobic patch that is exposed to the solvent. This patch is of functional significance that could potentially interact with the C-terminus of RNR. This hydrogen bonding network lends to the stability of the redox active site <ref name="Phulera" />. | ||
| + | |||
| + | A highly conserved residue in all members of the NrdH family is Arg-68. This residue hydrogen bonds with the main carbonyl oxygen of His-60. The His-60 is located before the WSGFRP motif, which suggests that the interaction between Arg-68 and His-60 may be of structural significance. In an alternate conformation of Arg-68, the guanidinyl group of Arg-68 forms a salt bridge with Asp-59. The hydrogen bonds and salt bridge work together to stabilize the WSGFRP motif <ref name="Phulera" />. | ||
| + | |||
| + | A buried water molecule binds with the WSGFRP motif. This water is believed to be one of the structural signatures of NrdH proteins. The hydrogen bonding network of the CVQC and WSGFRP motifs also involves the water molecule, and this may suggest that this region is important in the evolution of NrdH <ref name="Phulera" />. | ||
| + | |||
| + | == Function == | ||
| + | The main function is to act as a reducing partner of class 1B ribonucleotide reductase and for ribonucleotide reduction (RR), it is thought to supply electrons for this biochemical reaction. RR is one of the most fundamental biochemical processes that is required for DNA based life form to exist. Ribonucleotide reductases (RNRs) produce deoxyribonucleotides. These are precursors for DNA synthesis <ref name="Phulera" />. | ||
== Chemical Processes == | == Chemical Processes == | ||
| Line 41: | Line 50: | ||
Tuberculosis, also known as TB, is caused by ''Mycobacterium tuberculosis'' which normally attacks the lungs. TB bacteria attack the lungs, kidneys, spine, and the brain as well. This disease is fatal if not treated properly and was the leading cause of death in the United States in the past. It can be spread through the air from one person to another by coughing, sneezing, or speaking <ref>DOI: 10.1016/j.tube.2003.08.003</ref>. | Tuberculosis, also known as TB, is caused by ''Mycobacterium tuberculosis'' which normally attacks the lungs. TB bacteria attack the lungs, kidneys, spine, and the brain as well. This disease is fatal if not treated properly and was the leading cause of death in the United States in the past. It can be spread through the air from one person to another by coughing, sneezing, or speaking <ref>DOI: 10.1016/j.tube.2003.08.003</ref>. | ||
| - | Latent TB infections can live inside the human body in a dormant state and therefore will not get sick and will not be contagious. When the immune system cannot support the latent TB bacteria anymore, it will become active and contagious. This can be treated with several drugs taken for 6-9 months <ref>Centers for Disease Control and Prevention</ref> | + | Latent TB infections can live inside the human body in a dormant state and therefore will not get sick and will not be contagious. When the immune system cannot support the latent TB bacteria anymore, it will become active and contagious. This can be treated with several drugs taken for 6-9 months <ref>Centers for Disease Control and Prevention</ref>. |
| - | == Relevance == | ||
[[Image:Image_7_(1).png|300px|right|thumb|Sequence alignment of NrdH from ''Mycobacterium tuberculosis'', ''Corynebacterium glutamicum'', and ''Echerichia coli'']] | [[Image:Image_7_(1).png|300px|right|thumb|Sequence alignment of NrdH from ''Mycobacterium tuberculosis'', ''Corynebacterium glutamicum'', and ''Echerichia coli'']] | ||
| - | Genes that encode for NrdE and NrdF are essential for growth and RR might be an attractive biochemical pathway for antimycobacterial drug discovery. Organisms that depend solely on class 1B RNR could potentially | + | Genes that encode for NrdE and NrdF are essential for growth, and RR might be an attractive biochemical pathway for antimycobacterial drug discovery. Organisms that depend solely on class 1B RNR could potentially contain the essential genes and serve as potential drug targets for treating tuberculosis <ref name="Phulera" />. |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 03:38, 21 April 2015
Contents |
NrdH of Mycobacterium tuberculosis
Introduction
NrdH redoxins are small reductases, and they contain similar amino acid sequences to glutaredoxins and mycoredoxins. However, NrdH redoxins are different because of their thioredoxin-like activity. Their main function is to act as the electron donor for class 1b ribonucleotide reductases, which are important for the conversion of ribonucleotides to deoxyribonucleotides [1]. The process of ribonucleotide reduction is one of the most fundamental biochemical processes that is required for the existence of DNA-based life. It is the only de novo pathway to synthesize deoxyribonucleotides. Deoxyribonucleotides are the building blocks of DNA and are synthesized from ribonucleotides by reducing the 2’OH in a radical based reaction. The deoxyribonucleotides are then used as precursors for the process of DNA synthesis. This reaction is catalyzed by ribonucleotide reductases [2].
Ribonucleotide Reductases (RNRs)
All RNRs are related because they are necessary for all living cells, with the exception of a couple types of parasites and obligate endosymbionts. This is evident due to the catalytic core, which is structurally conserved across all extant RNRs. RNRs are essential for the processes of DNA replication and repair [3].
There are three classes of RNRs. These classes are divided based on the mechanism of radical generation in the reaction.
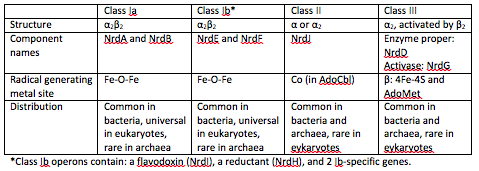
Class I enzymes reduce nucleotide 5’-diphosphates, while the other 2 classes reduce ribonucleotide 5’-triphosphates [2]. Class I RNRs are oxygen-dependent enzymes that contain a di-iron cluster [1].
Class 1 RNRs generate a tyrosyl radical in another subunit, which is NrdB for class 1a and NrdF in class 1b. The tyrosyl radical is then transferred to the catalytic subunit, which is NrdA in class Ia and NrdE in class 1b [3].
At the end of each cycle of ribonucleotide reduction, the ribonucleotide reductase needs to be reduced in order to be ready for the next reduction cycle. For a class 1a RNR, an external cofactor, such as a glutaredoxin or thioredoxin, performs this reduction step. For class 1b RNRs, this cofactor is known as NrdH. NrdH contains a glutaredoxin-like sequence but behaves like a thioredoxin [2].
NrdH of Mycobacterium tuberculosis
|
NrdH is a redox protein, and it is part of a family of redox proteins. The other proteins that maintain the redox balance of NrdH are three thioredoxin and three glutaredoxin-like proteins. Prokaryotes typically maintain redox homeostasis through low-molecular weight thiols (glutathione) and through proteins involved in disulfide exchange (thioredoxins).
NrdH is found in many types of bacteria, such as Mycobacterium tuberculosis. This bacteria causes the disease tuberculosis, which is a fatal disease if not treated properly. Tuberculosis was the leading cause of death in the United States in the past, and it can be spread through the air from one person to another by coughing, sneezing, or speaking [4].
Structure
The tertiary structure of NrdH has a thioredoxin fold with 79 residues with a glutaredoxin-like sequence. However, unlike glutaredoxins, NrdH of Mycobacterium tuberculosis can accept electrons from thioredoxin reductase. The binding site of NrdH is specific for aromatic amino acids [2].
Conserved Motifs
Members of the NrdH family are typically characterized by CVQC and WSGFRP
Within the , the amide oxygen of glutamine residue is firmly hydrogen bonded with the peptidyl nitrogen of Phe-44. The amide nitrogen of glutamine is then available for further hydrogen bonding. The carbonyl oxygen of Val-12 hydrogen bonds with peptidyl nitrogen of Ala-16. This hydrogen bonding leads to stability within the redox active site of NrdH. The N-terminal cysteine of the CVQC motif acts as a nucleophile, whereas the C-terminal cysteine acts as the resolving cysteine. The residues between the two cysteines are known to affect redox potentials and pKa values. Also, by changing the target proteins, in turn, they regulate the function [2].
The is stabilized by glutamine of the CVQC motif and phenylalanine is exposed to the solvent. Phe-64 and Val-12 with Ala-16 and Ala-20 create a distinct hydrophobic patch that is exposed to the solvent. This patch is of functional significance that could potentially interact with the C-terminus of RNR. This hydrogen bonding network lends to the stability of the redox active site [2].
A highly conserved residue in all members of the NrdH family is Arg-68. This residue hydrogen bonds with the main carbonyl oxygen of His-60. The His-60 is located before the WSGFRP motif, which suggests that the interaction between Arg-68 and His-60 may be of structural significance. In an alternate conformation of Arg-68, the guanidinyl group of Arg-68 forms a salt bridge with Asp-59. The hydrogen bonds and salt bridge work together to stabilize the WSGFRP motif [2].
A buried water molecule binds with the WSGFRP motif. This water is believed to be one of the structural signatures of NrdH proteins. The hydrogen bonding network of the CVQC and WSGFRP motifs also involves the water molecule, and this may suggest that this region is important in the evolution of NrdH [2].
Function
The main function is to act as a reducing partner of class 1B ribonucleotide reductase and for ribonucleotide reduction (RR), it is thought to supply electrons for this biochemical reaction. RR is one of the most fundamental biochemical processes that is required for DNA based life form to exist. Ribonucleotide reductases (RNRs) produce deoxyribonucleotides. These are precursors for DNA synthesis [2].
Chemical Processes
NrdH is able to accept electrons from M. tuberculosis thioredoxin reductase and is able to reduce the disulfide bonds that are present in insulin [2].
Disease
Tuberculosis, also known as TB, is caused by Mycobacterium tuberculosis which normally attacks the lungs. TB bacteria attack the lungs, kidneys, spine, and the brain as well. This disease is fatal if not treated properly and was the leading cause of death in the United States in the past. It can be spread through the air from one person to another by coughing, sneezing, or speaking [5].
Latent TB infections can live inside the human body in a dormant state and therefore will not get sick and will not be contagious. When the immune system cannot support the latent TB bacteria anymore, it will become active and contagious. This can be treated with several drugs taken for 6-9 months [6].
Genes that encode for NrdE and NrdF are essential for growth, and RR might be an attractive biochemical pathway for antimycobacterial drug discovery. Organisms that depend solely on class 1B RNR could potentially contain the essential genes and serve as potential drug targets for treating tuberculosis [2].
References
- ↑ 1.0 1.1 Van Laer K, Dziewulska AM, Fislage M, Wahni K, Hbeddou A, Collet JF, Versees W, Mateos LM, Tamu Dufe V, Messens J. NrdH-redoxin of Mycobacterium tuberculosis and Corynebacterium glutamicum dimerizes at high protein concentration and exclusively receives electrons from thioredoxin reductase. J Biol Chem. 2013 Jan 28. PMID:23362277 doi:http://dx.doi.org/10.1074/jbc.M112.392688
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Phulera S, Mande SC. The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87 A Suggests a Possible Mode of Its Activity. Biochemistry. 2013 May 28. PMID:23675692 doi:10.1021/bi400191z
- ↑ 3.0 3.1 Lundin D, Gribaldo S, Torrents E, Sjoberg BM, Poole AM. Ribonucleotide reduction - horizontal transfer of a required function spans all three domains. BMC Evol Biol. 2010 Dec 10;10:383. doi: 10.1186/1471-2148-10-383. PMID:21143941 doi:http://dx.doi.org/10.1186/1471-2148-10-383
- ↑ Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb). 2004;84(1-2):29-44. doi: 10.1016/j.tube.2003.08.003. PMID:14670344 doi:http://dx.doi.org/10.1016/j.tube.2003.08.003
- ↑ Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb). 2004;84(1-2):29-44. doi: 10.1016/j.tube.2003.08.003. PMID:14670344 doi:http://dx.doi.org/10.1016/j.tube.2003.08.003
- ↑ Centers for Disease Control and Prevention