Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
== Structural Highlights == | == Structural Highlights == | ||
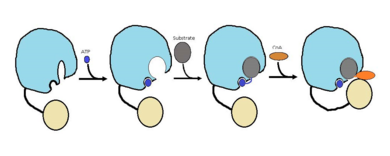
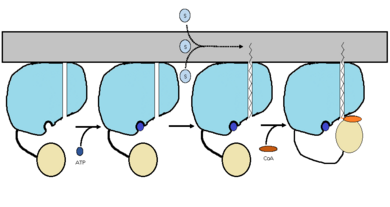
| - | FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a peripheral-membrane | + | FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a [http://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral-membrane protein]. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. There are two domains of this protein, a larger N-terminal domain (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and a smaller C-terminal domain (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>). These domains are held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>). Inside the larger N-terminal domain is a hydrophobic tunnel, which allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine-rich lid loop that is involved in the peripheral binding of the enzyme to the membrane<ref> [http://www.proteopedia.org/wiki/index.php/3r44/ 3R44] </ref>. Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell wall. |
== Function == | == Function == | ||
Revision as of 12:58, 14 April 2015
FadD13
| |||||||||||